Introduction:
Transcription factors are proteins with domains that bind to the DNA at the DNA-regulatory sequences (promoter or enhancer) of particular genes. Generally, the regulatory region is located in the 5’ upstream region of target genes, to modulate the rate of gene transcription, protein synthesis and subsequent altered cellular function. Additionally, they contain a domain that cooperates RNA polymerase II or other transcription factors, regulating the amount of messenger RNA (mRNA) that the gene is going to synthesised.
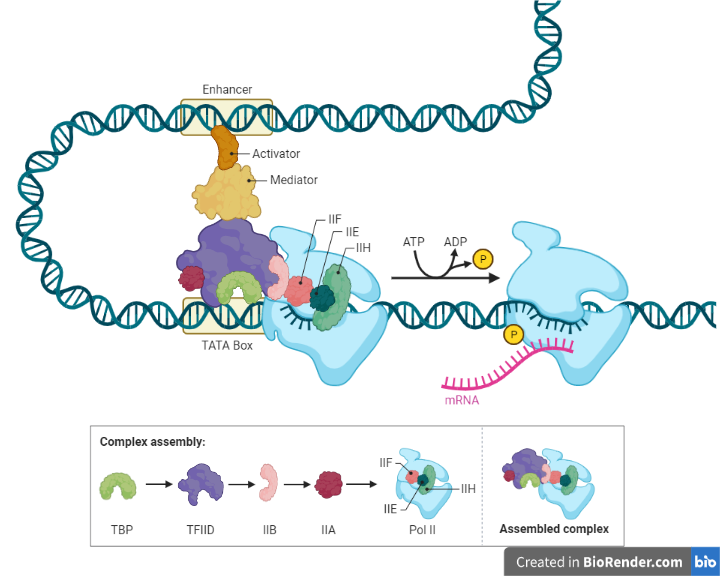
Fig: Eukaryotic Gene Regulation – Transcriptional Initiation
General transcription factors are a class of proteins that bind to a promoter on DNA to activate transcription. Nevertheless, without enhancers, the expression level remains only basal. An enhancer region in DNA recruit’s activators responsible for stabilizing promoter binding through DNA looping and interactions with a mediator. Unphosphorylated CTD of RNA Pol-II binds to mediators whereas phosphorylation of CTD release mediator and stimulates promoter release.
Eukaryotic and Prokaryotic transcription factors:
| Structural Feature | Eukaryotic Transcription Factors | Prokaryotic Transcription Factors |
| Size | Large, with multiple domains | Small, with one or two domains |
| DNA-Binding Domain | Commonly uses helix-turn-helix motifs or zinc fingers | Commonly uses helix-turn-helix motifs |
| Transcriptional Activation Domain | Interacts with co-activators to promote transcription | Short peptide sequence that interacts with RNA polymerase to facilitate transcription initiation |
| Repression Domain | Interacts with co-repressors to inhibit transcription | May lack a repression domain |
| Chromatin-Remodeling Domain | Can modify the structure of chromatin to regulate gene expression | Not typically present |
| Protein-Protein Interaction Domain | Allows interaction with other proteins involved in transcriptional regulation | May have a specific protein interaction domain |
| Additional Domains | May have protein kinase, DNA helicase, or other domains for further regulation | May have effector domains that bind small molecule ligands to regulate activity |
Transcription factors Binding Sites (TFBS):
The mechanism of transcriptional regulation depends on the binding of transcription factors to transcription factor binding sites (TFBSs). Transcription factors (TFs) recognize and bind to certain DNA sequences known as transcription factor binding sites (TFBS) in order to control the expression of genes. Variable distances from the transcription start site (TSS) are found in the promoter regions of genes where TFBS often contain conserved sequence patterns. Many genes are regulated by multiple TFs, frequently through combinatorial and cooperative binding mechanisms. The accessibility of TFBS is influenced by chromatin structure and modifications. Gene expression and TF binding can both be impacted by epigenetic changes. In general, understanding the characteristics of TFBS is crucial for comprehending gene regulation and how it is regulated by different cellular events.

Fig: Eukaryotic and Prokaryotic Gene Structure
How do transcription factors work?
Transcription factors are proteins that regulate gene expression by controlling the transcription of specific genes into RNA. They do this by binding to specific DNA sequences within the promoter or enhancer regions of target genes, either directly or through other proteins. This binding can either promote or inhibit the recruitment of RNA polymerase to the gene, which ultimately affects the transcription of the gene into RNA. Transcription factors can also interact with other proteins, such as co-activators or co-repressors, to further regulate gene expression. In addition to binding to DNA, transcription factors can also modify chromatin structure through interactions with histone proteins or enzymes that modify histones. This can alter the accessibility of the DNA to the transcriptional machinery, further regulating gene expression. Overall, transcription factors play a critical role in controlling gene expression and are essential for normal cellular processes such as cell growth, differentiation, and response to environmental cues.
The process by which transcription factors work can be summarized in the following steps:
- DNA binding:
- Recruitment of co-activators or co-repressors:
- Regulation of chromatin structure:
- Recruitment of RNA polymerase:
- Termination:
Transcription factors and disease:
In humans, dozens of transcription factors, cofactors, and chromatin regulators work together to regulate the gene expression programs that generate and maintain particular cell states. Numerous diseases can be caused on by the improper regulation of these gene expression.

Source: https://doi.org/10.1056/nejm199601043340108
Fig: Mechanism of Action of Normal and Mutated Transcription Factors That Result in mutation and diseases
The appropriate functioning of transcription factors is necessary for normal cellular functions because they are essential regulators of gene expression. Different diseases, such as cancer, metabolic disorders, neurological disorders, and autoimmune diseases, can be caused on by the dysregulation of transcription factors. In addition to post-translational modifications, protein-protein interactions, and DNA-binding interactions, transcription factors can either stimulate or inhibit the expression of genes. Understanding the function of transcription factors in disease can help identify prospective therapy targets as well as the pathophysiology of the illness. A number of medications that target transcription factors are presently undergoing clinical trials. Transcription factors have also been investigated as possible targets for therapeutic development.
Alteration of transcription factors:
The genes that encode transcription factors can become mutated, or they can undergo post-translational changes. These changes may have an impact on the transcription factor’s binding affinity, stability, localization, and activity, which may eventually result in modifications to gene expression. Changes in transcription factors have been connected to the emergence of a number of diseases, including cancer, diabetes, and neurological conditions. For these diseases to have targeted therapeutics, it is crucial to comprehend the mechanisms of transcription factor changes and how they affect gene expression.
Common transcription factors:
There are many different transcription factors that are involved in regulating gene expression in various organisms and cell types. These are just a few examples of the many transcription factors that are involved in regulating gene expression in different cell types and biological processes.
CREB (cAMP response element-binding protein): This transcription factor is involved in the regulation of genes that are activated by cAMP signaling, and plays a role in learning and memory.
NF-κB (nuclear factor kappa B): This transcription factor is involved in the regulation of immune responses and inflammation, and is activated in response to various stimuli, including infection and stress.
p53: This transcription factor is involved in the regulation of cell cycle and apoptosis, and is commonly mutated in many types of cancer.
AP-1 (activator protein-1): This transcription factor is involved in the regulation of cell proliferation, differentiation, and apoptosis, and is activated in response to various stimuli, including growth factors and stress.
HIF-1 (hypoxia-inducible factor 1): This transcription factor is involved in the response to low oxygen levels, and plays a role in angiogenesis and tumor growth.
MyoD: This transcription factor is involved in the regulation of muscle differentiation, and is required for the formation of skeletal muscle.
Sox2: This transcription factor is involved in the regulation of embryonic development, and is required for the maintenance of pluripotency in embryonic stem cells.
Oct4: This transcription factor is involved in the regulation of embryonic development and stem cell pluripotency, and is required for the maintenance of embryonic stem cells.
References:
- Latchman DS. Transcription factors: an overview. Int J Exp Pathol. 1993 Oct;74(5):417-22. PMID: 8217775; PMCID: PMC2002184.
- Vinson C, Chatterjee R, Fitzgerald P. Transcription factor binding sites and other features in human and Drosophila proximal promoters. Subcell Biochem. 2011; 52:205-22.
- Pulverer, B. Sequence-specific DNA-binding transcription factors. Nature Milestones (2005)
- Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013 Mar 14;152(6):1237-51
