Background:
In this recent world, amplification and detection of nucleic acids are among the most important techniques employed in biological research and study. These approaches are used by scientists in a variety of fields, including basic science, biotechnology, medicine, forensic science, diagnostics, and others.
Introduction:
- A real-time polymerase chain reaction (RT- PCR) which is also known as qPCR is a molecular biology laboratory technique that is based on the polymerase chain reaction (PCR).
- It is rapidly becoming the “gold standard” for detecting and quantifying nucleic acid sequences.
- It monitors the amplification of a specific DNA molecule during the PCR (rather than at the end, as in conventional PCR).
- The amplified product, or amplicon, is detected in conventional PCR by running DNA on an agarose gel electrophoresis once the process has ended.
- Real-time PCR, on the other hand, allows the buildup of amplified product to be observed and evaluated as the reaction occurs, i.e., in “real time.” Real-time detection of PCR products is made possible by introducing a fluorescent molecule in the reaction that signals an increase in DNA amount with a corresponding increase in fluorescence signal.
- DNA-binding dyes and fluorescently labeled sequence-specific primers or probes are among the fluorescent technologies used for this purpose.
- Fluorescence is monitored while amplification occurs using specialized thermo cycler equipped with fluorescence detector modules. The amount of amplified product in each cycle is reflected in the observed fluorescence.
Principle:
In RT-PCR, the RNA template is first transformed into complementary DNA (cDNA) using a reverse transcriptase (RT). The cDNA is subsequently utilized as a template for PCR exponential amplification by Polymerase chain reaction (PCR).
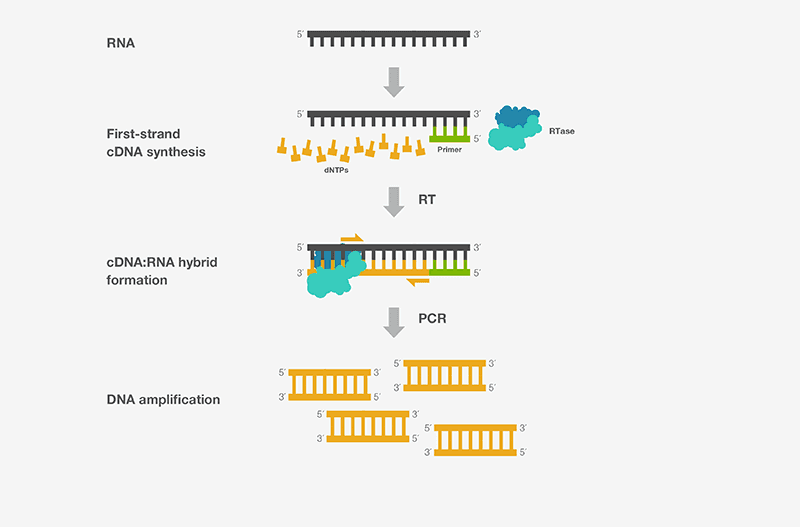
Fig: Principle of RT-PCR
Components:
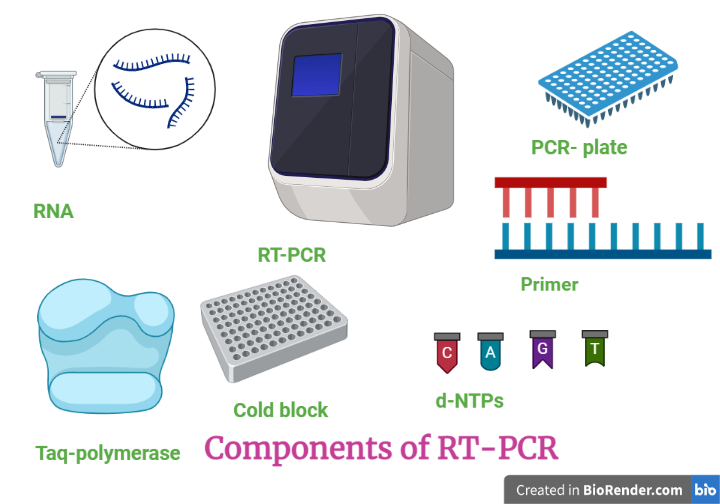
Fig: Component required in RT-PCR
Procedure:
Basically, it is dived in two different sections.
Amplification
Denaturation
High temperature incubation is used to “melt” double-stranded DNA into single strands and release secondary structure in single-stranded DNA. The maximum temperature that the DNA polymerase can tolerate is usually employed (normally 95°C).
Annealing
In this step, the temperature is reduced in order for the DNA primers to bind to the template DNA.
Extension
At 70-72°C, the activity of the DNA polymerase is optimal, and primer extension occurs at rates of up to 100 bases per second.

Fig: Steps of RT-PCR
Detection
Two common methods for the detection of PCR products in real-time PCR are
(1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and
(2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence.
SYBR Green and Taqman are two technologies used to detect or monitor real-time PCR amplification.
Both technologies are designed to generate fluorescence during the PCR, which allows real-time PCR machine to monitor the reaction in “real time”. However, there is the key difference between SYBR Green and Taqman
- SYBR Green method is carried out using a fluorescent dye called SYBR green and detects the amplification by binding the dye to produced double stranded DNA.
- Taqman is carried out using dual-labeled probes and detects the amplification by degradation of the probe by Taq polymerase and releases of the fluorophore.
SYBR Green
SYBR Green is a fluorescent dye used in molecular biology to stain nucleic acids, particularly double-stranded DNA.
During real-time PCR, the SYBR Green technique is utilized to quantify PCR products. The resultant DNA-dye complex absorbs blue light and emits strong green light after binding with DNA. It occurs as a result of the structural change that occurs in the dye molecule when it binds to double-stranded DNA.
As PCR generates more and more DNA, more dye molecules link to it, resulting in increased fluorescence. As a result, the fluorescence increases as the PCR product accumulates. Consequently, the amount of PCR product can be quantified using SYBR Green fluorescence detection.
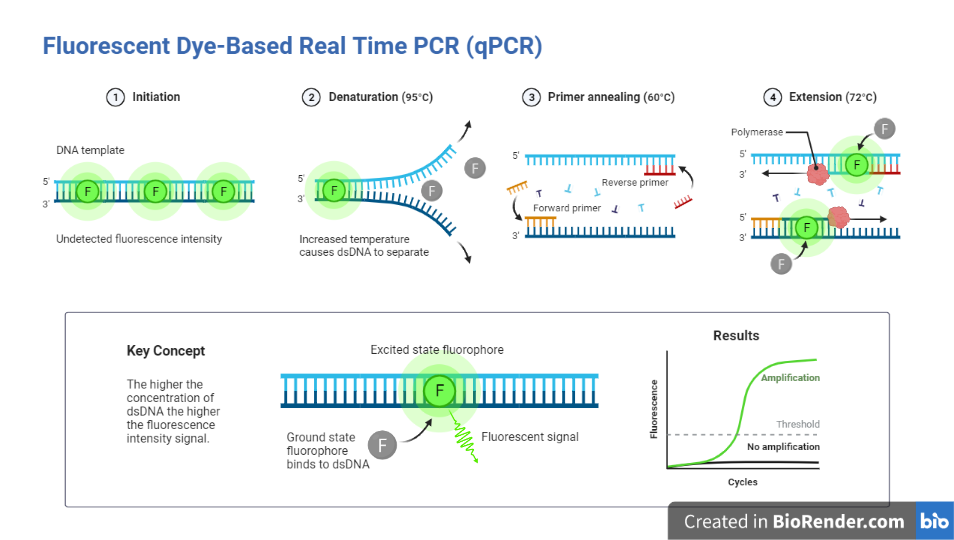
Fig: Fluorescent dye-based PCR
Taqman
It is an alternative to SYBR Green for monitoring the real-time PCR process.
This approach relies on 5′ – 3′ exonuclease activity of the Taq polymerase enzyme to destroy probes during new strand extension and fluorophore release. This approach, which is based on probe hydrolysis, employs dual-labeled probes. Fluorescently labeled DNA oligonucleotides with a fluorescent reporter molecule (fluorophore) at the 5′ end and a quencher molecule at the 3′ end are known as probes.
They are intended to bind to the single-stranded template on the other side of the primer anneals.
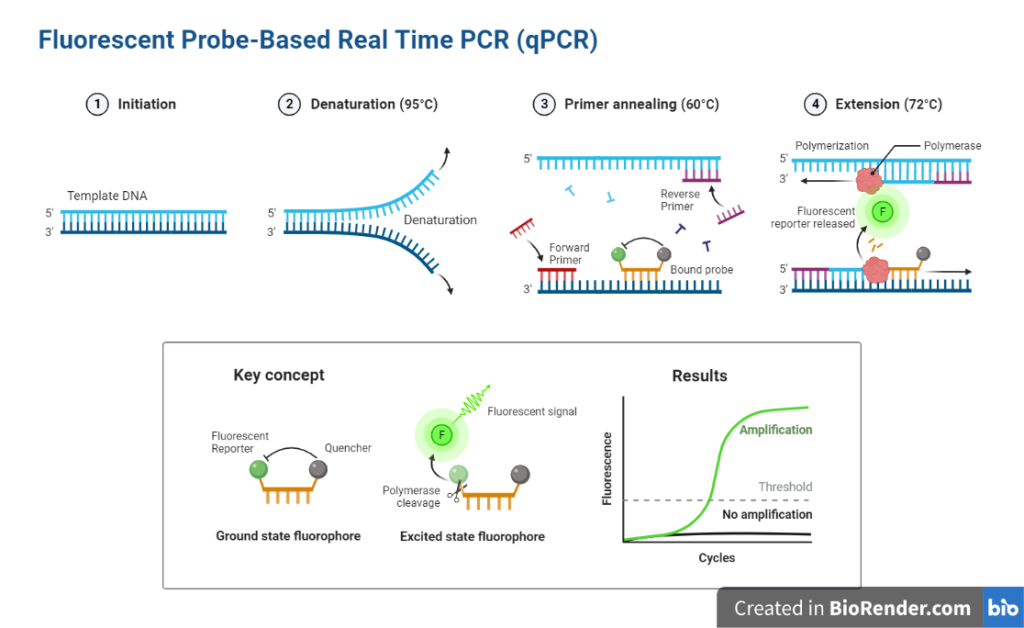
Fig: Fluorescent probe-based PCR
Taq polymerase adds nucleotides to the primer and stretches the new strand towards the dual-labeled probes.
Taq polymerase exonuclease activity activates and destroys the probe once it comes into contact with it.
After completing the synthesis of the new strand, the probe is subjected to complete breakdown, releasing the fluorophore.
The fluorophore release generates fluorescence, which is used to quantify the PCR result.
The release of fluorophores and the number of PCR products are proportional. As a result, the Taqman approach simplifies quantification.
Types:
When RNA is employed as the starting material, quantitative reverse transcription PCR (RT-qPCR) is used. In this process, reverse transcriptase converts total RNA or messenger RNA into complementary DNA (cDNA) (mRNA). Thereafter, the cDNA is employed as a template for the qPCR reaction.
One-step RT-PCR
In a single reaction mixture with a common reaction buffer, cDNA synthesis and PCR are carried out. The reverse transcriptase and DNA polymerase are mixed thoroughly into a single tube in the one-step method. This enables the RT and subsequent amplification steps to be carried out in a single reaction.
Gene-specific primers govern the synthesis and amplifying of a particular target’s cDNA. One-step reactions provide many benefits, including as less sample handling, quicker bench times, and closed-tube reactions that lessen the possibility of cross-contamination and pipetting errors.
The efficiency of one-step RT-PCR is affected by the quality and availability of RNA samples. Because the product of cDNA synthesis cannot be stored after one-step RT-PCR, additional aliquots of the original RNA sample(s) are necessary to repeat reactions or analyze the expression of other genes.
Two-step RT-PCR
In two-step RT-PCR, the reverse transcription of the RNA template is performed first. Once completed, the amplification of the cDNA is carried out in a separate reaction. cDNAs thus synthesized are amplified using specific primers in a subsequent PCR experiment.
This requires extra open-tube step, more pipetting manipulations, and longer hands-on time which may lead to greater variability and risk of contamination.
The remaining cDNA can be preserved for future use, or the expression of multiple genes can be quantified from a single RNA/cDNA sample.
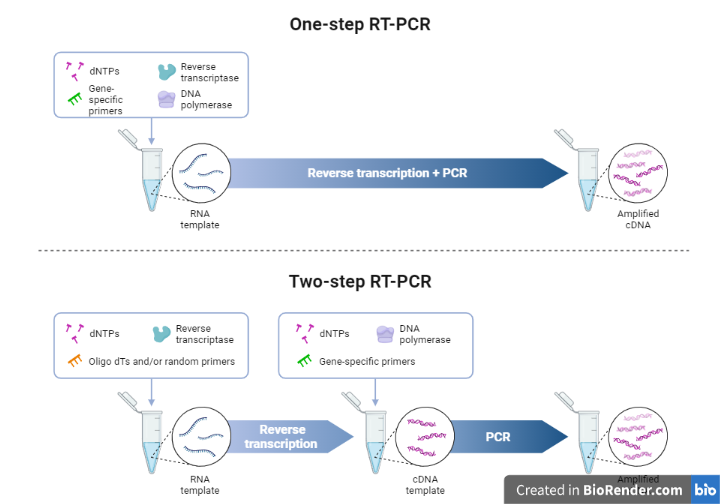
Fig: One step and two steps PCR
Advantages:
- The fundamental advantage of real-time PCR over conventional PCR is that it allows to accurately and sensitively identify the beginning template copy number over a large dynamic range.
- The results of real-time PCR might be either qualitative (the presence or absence of a sequence) or quantitative (number of copies of DNA).
- Furthermore, real-time PCR data can be analyzed without using gel electrophoresis, resulting in shorter experiment times and higher throughput.
- Reactions are performed and data is analyzed in a closed-tube system that will reduce contamination and eliminate postamplification manipulation.
Applications:
- Gene expression analysis, RNAi validation, microarray validation, pathogen identification, genetic testing, and clinical research are all applications for RT-qPCR.
- Real-time PCR (quantitative PCR, qPCR) is currently a well-established technology for detecting, quantifying, and typing many microbial agents in clinical and veterinary diagnostics, as well as food safety.
- It is used to identify genetically engineered organisms (GMOs)
- RT-PCR is commonly used to detect expressed genes, examine transcript variations, and generate cDNA templates for cloning and sequencing.
- It is a simple and inexpensive technique for determining target gene expression levels that is commonly utilized in biomedical science research, including nanotoxicology studies, for semiquantitative analysis.
References:
- Edwards, K., Logan, J. and Saunders, N., 2004. Real-time PCR: an essential guide. Horizon Bioscience.
- Bio-Rad Laboratories, 2006. Real-time PCR applications guide. Bulletin 5279, pp.2-6.
- https://www.thermofisher.com/np/en/home/life-science/pcr/real-time-pcr/real-time-pcr-learning-center/real-time-pcr-basics/essentials-real-time-pcr.html
- Molecular Cloning and Recombinant DNA Technology. Matt Carter, Jennifer Shieh, in Guide to Research Techniques in Neuroscience (Second Edition), 2015
- Kaltenboeck, B. and Wang, C., 2005. Advances in real‐time PCR: Application to clinical laboratory diagnostics. Advances in clinical chemistry, 40, p.219.
- Bustin S. (ed) (2004) A-Z of Quantitative PCR. IUL Biotechnology Series, International University Line, La Jolla, California.
- Mo Y, Wan R, Zhang Q. Application of reverse transcription-PCR and real-time PCR in nanotoxicity research. Methods Mol Biol. 2012; 926:99-112. doi: 10.1007/978-1-62703-002-1_7. PMID: 22975959; PMCID: PMC5087796.
