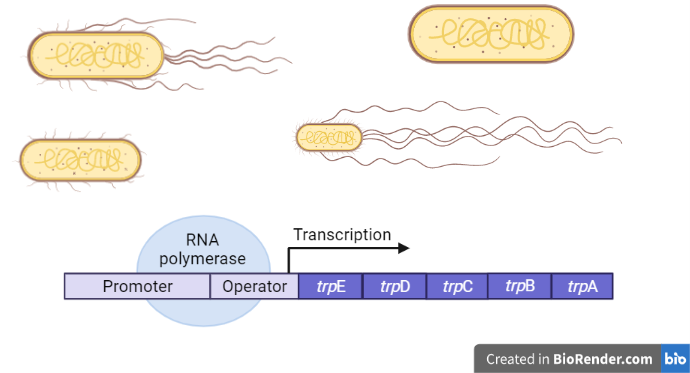
Background:
Bacteria like E. coli require amino acids to exist because they, like us, must produce proteins. Tryptophan is an essential amino acid for them. E. coli will take up tryptophan if it is present in the environment and use it to make proteins. E. coli, on the other hand, can also synthesis tryptophan on their own if needed utilizing enzymes encoded by five genes. These five genes, which are located in close proximity to each other, are found in the trp operon. It is an example of anabolic operon.
A group of genes transcribed under the regulation of an operator gene is known as an operon. An operon, in more detail, is a DNA segment that contains adjacent genes such as structural genes, operator genes, and regulatory genes. Thus, an operon is a transcriptional and genetic regulatory functional unit.
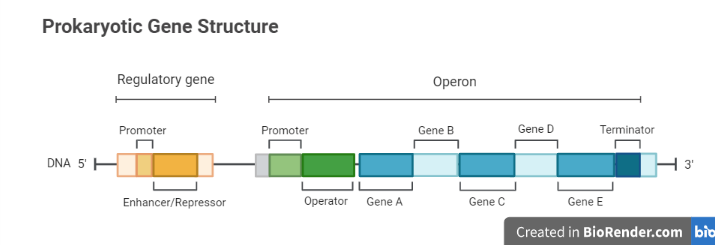
Fig: Structrue of prokaryotic Gene
The trp operon was first discovered in Escherichia coli, however it can be found in numerous species. Operons are bacterial genome features that are made up of groups of co-regulated genes with related functions. The lac operon in Escherichia coli was the first classical operon to be defined, and Jacob and Monod invented the term “operon.”
Introduction:
The tryptophan operon is a set of genes that code for tryptophan-producing enzymes. E. coli’s trp operon contains five important structural genes that encode all seven protein functional domains required for tryptophan biosynthesis from chorismate, a common aromatic precursor. The trp operon’s transcription is tightly controlled.
When tryptophan is present in the environment, the operon is controlled to prevent the expression of tryptophan synthesis genes. It was a crucial experimental way of learning about gene expression, and it’s still addressed today.
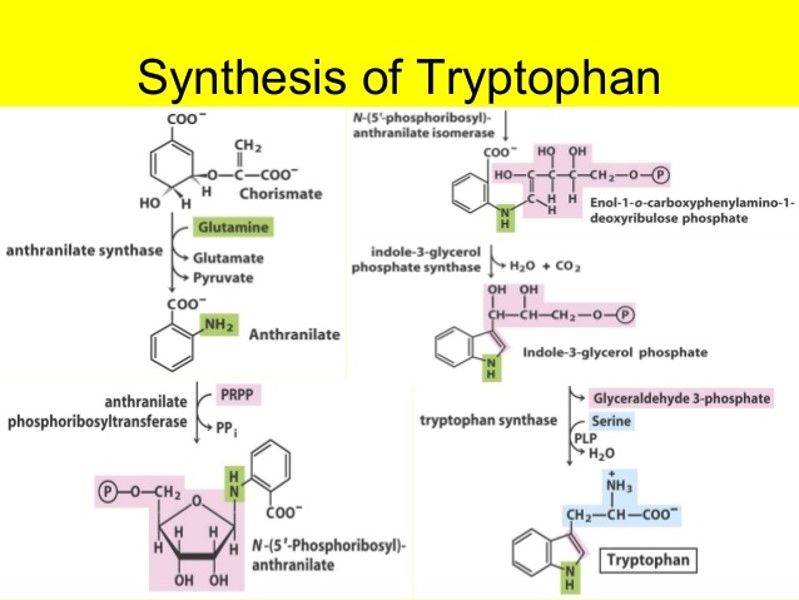
Fig: Pathway for the synthesis of tryptophan
These enzymes are translated in E. coli from a single polycistronic mRNA. A promoter, an operator, and two sections known as the leader and the attenuator are located adjacent to the enzyme coding sequences in the DNA. The sequences of the leader and attenuator are transcribed. A repressor gene (trpR) is positioned some distance away from this gene cluster.
trpR has a promoter where RNA polymerase binds and synthesizes mRNA for a regulatory protein. The protein that is synthesized by trpR then binds to the operator which then causes the transcription to be blocked. In the trp operon, tryptophan binds to the repressor protein effectively blocking gene transcription.
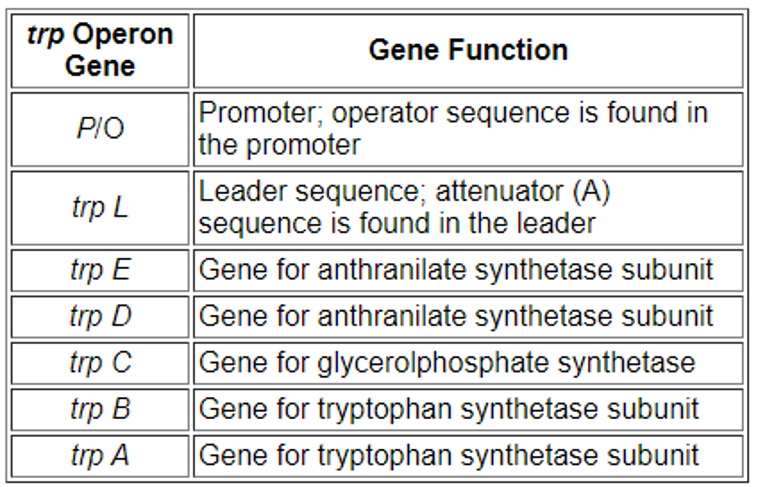
Fig: Function of different gene in tryptophan operon
Regulation:
Repression
When tryptophan is high in cell
When tryptophan is high in cell then it binds with repressor protein and change its confirmation so that it become active and bind to the operator near promoter.
The repressor protein’s binding to the operator overlaps the promoter, preventing RNA polymerase from binding to it.
Since tryptophan is already high in cell, no transcription of structural gene is required for biosynthesis of tryptophan. This is also known as negative regulation.
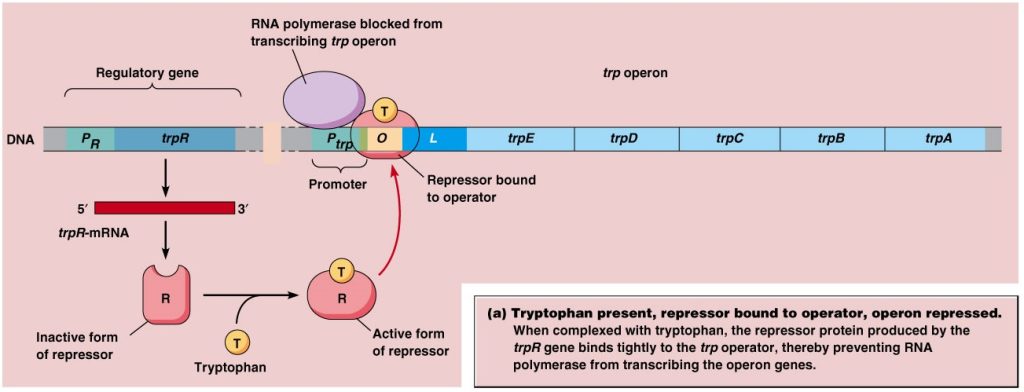
Fig: When tryptophan is high in cell
When tryptophan is absent in cell
The repressor protein which is encoded by repressor gene (trpR) that is inactive. In the absence of tryptophan, transcription of structural gene occurs for the biosynthesis of tryptophan from chorismate.
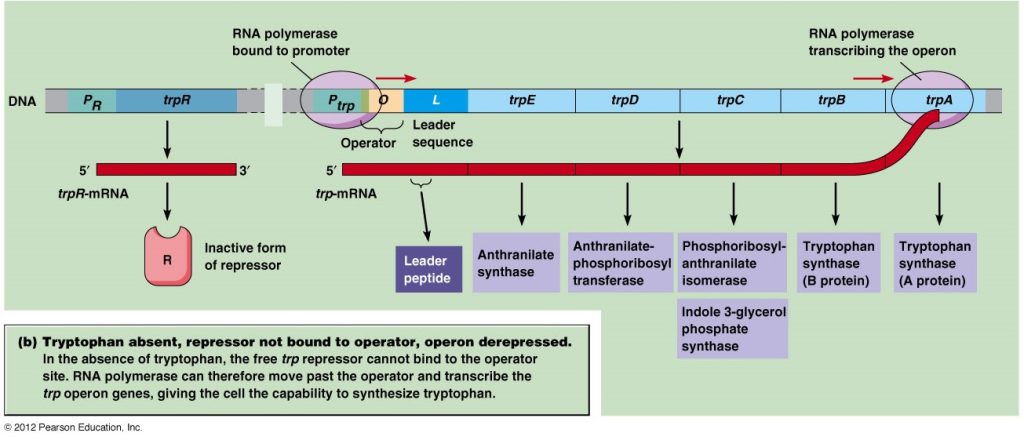
Fig: When tryptophan is absent in cell
Attenuation
In addition, the elongating transcription complex is sensitive to attenuation regulation once transcription has begun. Repression (80-fold) and attenuation (8-fold) work together to give the trp operon 600-fold control over transcription in response to different levels of tryptophan availability.
Once repression is lifted and transcription begins, the rate of transcription is fine-tuned by a second regulatory process, called transcription attenuation, in which transcription is initiated normally but is abruptly halted before the operon genes are transcribed. The frequency with which transcription is attenuated is regulated by the availability of tryptophan and relies on the very close coupling of transcription and translation in bacteria.
The trp operon attenuation mechanism uses signals encoded in four sequences within a 162-nucleotide leader region at the 5, end of the mRNA, preceding the initiation codon of the first gene. The attenuator, which is made up of sequences 3 and 4, is located within the leader.
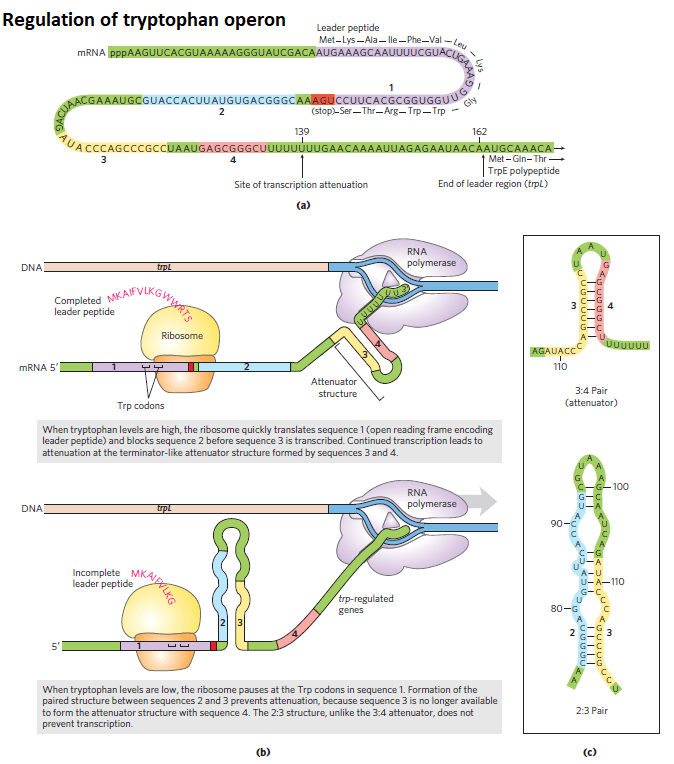
Fig: Trptophan control by attenuation
Transcription begins at the start of the 162-nucleotide mRNA leader, which is encoded by the trpL DNA region. Whether transcription is reduced at the end of the leader or continues into the structural genes is determined by a regulatory mechanism.
(a) The trp mRNA leader (trpL). The attenuation mechanism in the trp operon involves sequences 1 to 4.
(b) Sequence 1 encodes a small peptide, the leader peptide, containing two Trp residues (W); it is translated immediately after transcription begins. Sequences 2 and 3 are complementary, as are sequences 3 and 4. The attenuator structure forms by the pairing of sequences 3 and 4. Its structure and function are similar to those of a transcription terminator. Pairing of sequences 2 and 3 prevents the attenuator structure from forming. It is be noted that the leader peptide has no other cellular function. Translation of its open reading frame has a purely regulatory role that determines which complementary sequences (2 and 3 or 3 and 4) are paired.
(c) Base-pairing schemes for the complementary regions of the trp mRNA leader.
Key points:
- In E. coli, the trp operon is present. coli bacterium, is a set of genes that code for tryptophan biosynthesis enzymes.
- When tryptophan levels are low, the trp operon is expressed (turned “on”), while when they are high, it is repressed (turned “off”).
- The trp repressor controls the expression of the trp operon. The trp repressor, when linked to tryptophan, prevents the operon from being expressed.
- Attenuation regulates tryptophan production as well (a mechanism based on coupling of transcription and translation).
References:
- Jacob F, Perrin D, Sanchez C, Monod J (1960) L’operon: Groupe de genes a l’expression coordonne par un operateur. C R Acad Sci 245: 1727–729 [PubMed]
- Jacob F, Monod J (1961) On the regulation of gene activity. In: Cold Spring Harbor Symposium Quantitative Biology 26, pp 193–211
- Platt, T., 1981. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell, 24(1), pp.10-23.
- Gu, P., Yang, F., Kang, J., Wang, Q. and Qi, Q., 2012. One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microbial cell factories, 11(1), pp.1-9.
- Nelson, D.L., Lehninger, A.L. and Cox, M.M., 2008. Lehninger principles of biochemistry. Macmillan.