Introduction:
- Blotting techniques involve the separation and transfer of DNA, RNA, or proteins onto a blotting membrane. It is very widely used analytical tools for the specific identification of desired DNA or RNA fragments from thousands of molecules This separation is generally followed by complexing of the target with a labelled molecule for detection.
- Blotting refers to the process of immobilization of sample nucleic acids on solid support (nitrocellulose or nylon membranes). The nucleic acids that have been blotted are subsequently employed as targets in hybridization assays to identify them specifically.
- Since Sir Southern described DNA blotting, now known as Southern blotting, blotting techniques have been an important aspect of life sciences research. Blotting is an investigative technique used to detect and identify macromolecules, such as nucleic acids and proteins.
General principle:
The blotting methods are fairly simple and usually consist of different steps that follow a general procedure:
- Isolation of target molecule in a sample
- Separation using electrophoresis
- Transferred (blotted) onto a filter or membrane
- Detection (Visualisation) with a labelled probe through expose to the membrane
Firstly, the sample will be separated by electrophoresis which is followed by transferring on to support medium or membrane. This immobilizes the protein or DNA fragments, allowing for a more accurate replication of the original separation and better biochemical analysis. The immobilized protein or nucleic acid fragment is localized that has been introduced to the support medium by using probes, such as antibodies or DNA, that precisely attach to the molecule of interest. Finally, colorant staining such as silver staining of proteins or by autoradiography is used to visualize the position of the probe attached to the immobilized target molecule.
Types of blotting:
All of these blotting procedures involve transferring molecules from a gel to a porous membrane, which is usually done by soaking a solution through the gel and then attaching absorptive paper to the membrane.
- Southern blotting is a technique for detecting certain DNA sequences that can be utilized in forensics and the identification of genetic mutations.
- Northern blotting is a technique for analysing gene expression primarily focuses on RNA sequences.
- Western blotting is a technique for detecting proteins and antibodies that can be used to diagnose viral disorders, protein abnormalities (like prion disease), and autoimmune diseases.
Carbohydrate epitopes, including glycoconjugates and lipids, are detected and analysed utilizing Eastern blotting.
Southern Blotting:
- Southern blotting is named after Edward M. Southern, a British scientist who developed the technique in the 1970s at Edinburgh University.
- It was the first method for detecting a particular DNA sequence in a variety of DNA samples. It combines electrophoresis-separated DNA fragment transfer to a filter membrane with probe hybridization enabling fragment detection.
- The identity, size, and quantity of DNA can be determined via Southern blot analysis. It’s a standard approach that requires electrophoresis to separate DNA fragments by size, transferring them to a membrane, hybridization with a labelled sequence-specific probe, washing, and finally detection of the labelled DNA band (s).
Steps:
DNA digestion
To extract the DNA from a cell’s nucleus, one must first lyse the entire cell to allow the DNA to be expelled. The entire cell is lysed when the cell culture is incubated with detergent. The proteinase enzyme is used to lyse the protein, which is then incubated. DNA, protein, and debris are all present in the lysed sample. Alcohol precipitation is used to purify and separate DNA, while a buffer is used to remove fibres. In Southern blot analysis, full fragmentation of the collected DNA at the desired restriction enzyme sites is crucial.
Gel electrophoresis
The molecular weights of fragmented DNA are separated by electrophoresis on an agarose gel. For better resolution of smaller DNA fragments (800 bp), acrylamide gels can be applied alternately.
A wide range of DNA ladders, including 100 bp ladders and 1 Kb Plus ladders, are available for accurate size and mass estimations for determining DNA size. The use of weak HCl for partial depurination increases improved efficiency transfer of DNA fragments by breaking them down into smaller fragments.
Denaturation
DNA thus attained are double stranded in nature. For the purpose of probe hybridization, single stranded DNA is required. DNA is therefore denatured in an alkaline solution which results in the formation of denatured DNA.
Blotting
Blotting is the process of transferring fragmented DNA sequences to nitrocellulose or nylon membranes. Electroblotting or capillary blotting are generally used to during the process. A NaCl solution is used to saturate the DNA molecule, which is then permanently fixed using UV light or drying. DNA being a negatively charged biomolecule is always transferred to a positively charged nylon membrane in electrophoresis. An upward-transfer approach is used to transfer DNA traditionally.
Baking
The nitrocellulose membrane is recovered from the blotting stack and baked in a vacuum or conventional oven at 80 °C for 2-3 hours or subjected to UV light to permanently bond the transferred DNA to the membrane.
Probe labelling
A nucleic acid probe with sequence homologous to the target sequence under study is labelled with radioactivity, fluorescent dye, or an enzyme that can generate a chemiluminescent signal when incubated with the appropriate substrate. The label chosen is determined by a number of parameters, including the nature of the probe or probe template, the required sensitivity, quantification parameters, convenience of use, and experimental time.
Hybridization & washing
During hybridization, the labelled probe is incubated with the DNA fragments that are immobilized on the blot under conditions that promote hybridization of complementary sequences. When used for both prehybridization and hybridization, hybridization buffer, can be employed that increase sensitivity
The unhybridized probe is then removed by washing it several times in different buffers. The hybridization solution and unhybridized probe are removed with low stringency washes. The result is that only fully hybridized labelled probe molecules, with complementary sequence to the region of interest, remain bound.
Detection
The bound, tagged probe is identified in the detection stage using the method required for the specific label utilized. For example, radiolabelled probes may be detected using X-ray film or a phosphorimaging instrument, and enzymatically labelled probes are typically detected by incubating with a chemiluminescent substrate and exposing the blot to X-ray film.

Fig: An outline of southern blotting
Transfer Methods:
There are three different alternative methods employed by which denatured DNA is transferred to the membrane such as;
Capillary Transfer (Wet or Tank Transfer)
In this approach, the transport of denatured DNA from the gel to the membrane is aided by the capillary flow of buffer from the wet paper to the dry paper.
Electrophoretic Transfer (Electroblotting)
In this process, the DNA is moved from the gel to the membrane with the help of an electric current.
Electrophoretic transfer is the most frequent method of transfer in southern blotting, in which DNA is eluted from gels and transferred to membranes using an electric field. The membrane and gel are joined together during this procedure, with filter paper sandwiched between two electrodes.
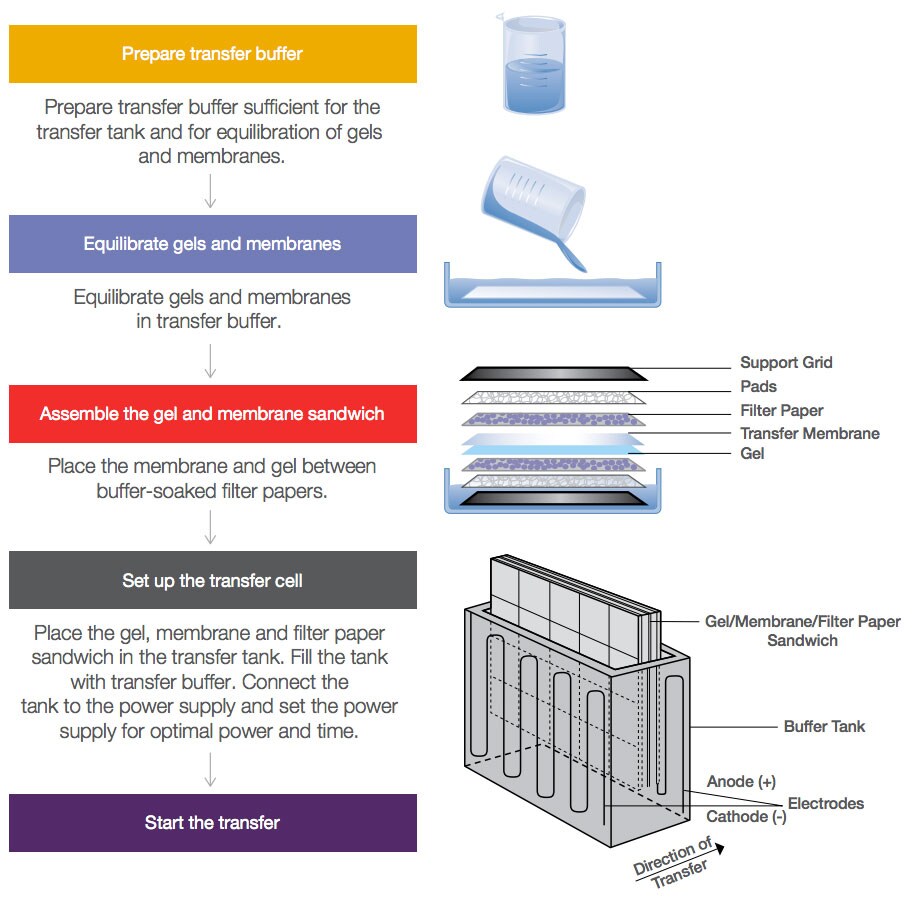
Fig: Transfer method in blotting technique
Vacuum Transfer
This approach uses suction to help transport the DNA from the gel to the membrane.
Detection method:
The main techniques for visualizing a western blot are colorimetric, chemiluminescence, and fluorescence.
Colorimetric and chemiluminescence act by an enzymatic reaction either by horseradish peroxidase or alkaline phosphatase (also used in ELISA). Meanwhile, fluorescence methods do not depend on a reaction but instead rely on fluorophore-conjugated antibodies that can be visualized directly.
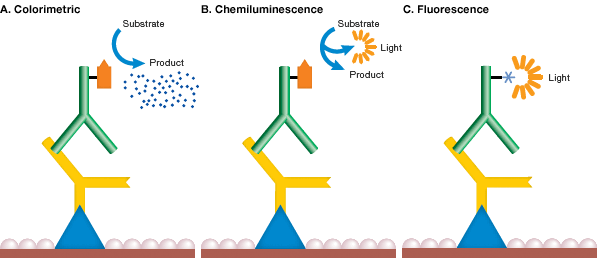
Fig: Detection methods in blotting
Applications:
- Southern blotting is mostly used to identify a specific DNA in a DNA sample.
- It is mostly used to diagnose viral infections as well as some bacterial diseases.
- DNA identification is utilized in evolutionary investigations, paternity and maternity analyses, forensic studies, and personal identification because of its precision.
- Southern blotting can be used to investigate a gene’s structure or to estimate restriction enzyme maps.
- Southern blotting’s identification of RFLPs aided in the mapping of numerous important genomes.
- Southern blotting can be used to examine clonal rearrangements of immunoglobulins and T Cell receptor genes, both of which are critical in generating an immunological response.
- It is utilized in rDNA technology (Recombinant DNA Technology) to isolate a specific DNA.
- It’s also significant in the research of mutations and gene rearrangements; this property is helpful to diagnose neonatal and genetic diseases.
References:
- Mitschka, S., Mayr, C. (2020). Northern blot protocol. Retrieved December 29, 2021, from https://www.protocols.io/view/northern-blot-protocol-bqqymvxw
- Tani Y, et al. Evaluation of a Novel Automated Machine, the Auto2D, for Two-Dimensional Gel Electrophoresis. J Proteomics Bioinform. 2014; 7:108-111.
- Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B., Alexander, J. S. (1997). An improved, rapid Northern protocol. Biochem Biophys Res Comm 238:277–279.
- Doustjalali SR, et al. Two-Dimensional Gel Electrophoresis: An Overview of Proteomic Technique in Cancer Research. J Proteomics Bioinform. 2014; 7:077-081.
