Introduction:
It is an invitro technique used to mutate one or more bases within a plasmid. Likewise, this procedure is to create specific, targeted changes in double stranded plasmid DNA with modified sequences for examining the importance of specific residues in protein structure and function. It is also called site-specific mutagenesis or oligonucleotide-directed mutagenesis. There are many reasons to make specific DNA alterations that includes insertions, deletions and substitutions.
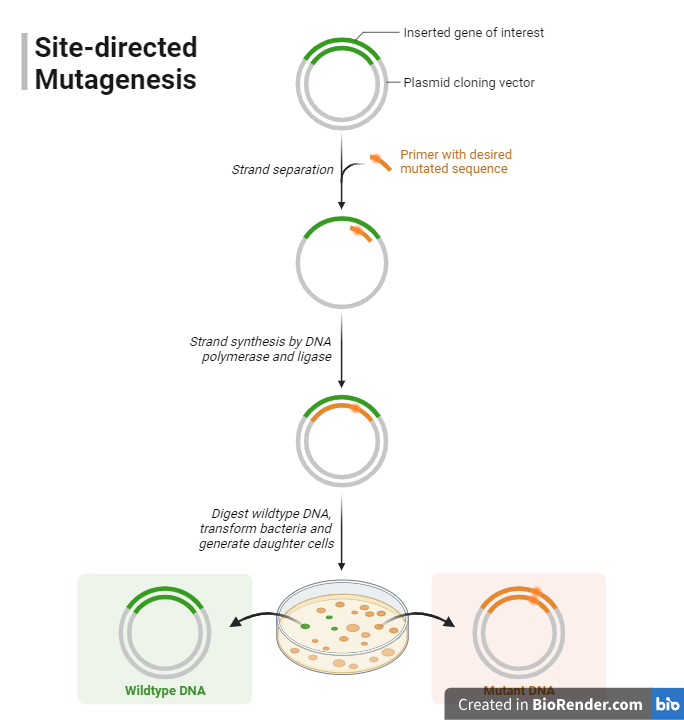
Fig: Site-Directed Mutagenesis (SDM)
Methods:
Basically, three types of site-directed mutagenesis are known.
Oligonucleotide-Directed Mutagenesis:
In this method, short, synthetic oligonucleotides containing the desired mutation(s) will be used as primers in a PCR reaction. These primers anneal to the template DNA, and during DNA synthesis, the mutation(s) will be incorporated into the newly synthesized DNA strands. It is relatively very simple and is appropriate for introducing point mutations or small insertions/deletions.
Inverse PCR-Based Mutagenesis:
This approach entails amplifying the complete plasmid or DNA fragment carrying the desired gene. After amplification, a restriction enzyme is used to linearize the DNA in the PCR product, and then self-ligation is performed. The intended mutations, which are inserted during the PCR amplification process, are present in the circularized DNA. Larger deletions or insertions as well as more complicated alterations can be produced via inverse PCR-based mutagenesis.
PCR-Based Site-Directed Mutagenesis:
This method, which involves producing mutagenic primers that carry the desired mutations, is the most popular. In a PCR reaction, these primers are combined with the template DNA. Following amplification, the original template DNA is removed via digestion with a restriction enzyme like DpnI, which focuses on methylated DNA. The unused PCR product is subsequently converted into a host organism for expression after containing the required mutations. Site-directed mutagenesis using PCR is flexible and effective for a variety of mutation types, including point mutations, deletions, and insertions.
The method of mutagenesis has been extremely successful, as witnessed by the growth of molecular biology and functional genomics, but suffers from a number of disadvantages.
- First, any gene in the organism can be mutated and the frequency with which mutants occur in the gene of interest can be very low.
- Second, even though the mutants with the desired phenotype will be isolated, there will be no assurance that the mutation has occurred in the specific gene of interest.
- Third, prior to the development of gene-cloning and sequencing techniques, there was no way of knowing where in the gene the mutation had occurred and whether it arose by a single base change, an insertion of DNA, or a deletion.
Polymerase Chain Reactions (PCR)-Based Site-Directed Mutagenesis:
Site-directed mutagenesis is a method that enables the introduction of particular mutations into a DNA sequence using the Polymerase Chain Reaction (PCR). This technique is frequently used in molecular biology to make specific alterations in a gene of interest.

Source: https://bitesizebio.com/
Fig: Protocol for inverse PCR in SDMPCR-based site-directed mutagenesis involves several key reactions to introduce specific mutations into a DNA sequence
- Primer Design: The first step is designing mutagenic primers that should contain the desired mutations and are essential for specifying the changes to make to the DNA sequence.
- PCR Amplification: It is used to amplify the DNA containing the desired mutation. This reaction includes denaturation, annealing of primers to the template DNA, and extension to create new DNA strands with the mutations.
- Digestion: After completion of PCR, digestion using restriction enzymes DpnI is performed. It specifically cleaves methylated DNA, such as the template DNA. This step eliminates the template DNA and retains the newly synthesized mutated DNA strands.
- Transformation: The mutated DNA generated will be transformed into a suitable bacterial host, often E. coli. This transformation reaction allows the mutated DNA to be incorporated into the genome of host.
- Selection and Screening: Colonies of transformed bacteria are selected by means of the selection method such as antibiotic selection which is used to isolate entities with specific traits. Additionally, confirmation of genetic modifications can be achieved through DNA sequencing, a pivotal step in genetic engineering experiments.
Applications:
- To study changes in protein activity that occur as a result of the DNA manipulation.
- To select or screen for mutations (at the DNA, RNA or protein level) that have a desired property
- To introduce or remove restriction endonuclease sites or tags
Site-Directed Mutagenesis and strain improvement:
Site-directed mutagenesis represents a modern and precise approach to microbial strain improvement, facilitating the targeted modification of microbial genomes for various beneficial purposes. This technique is instrumental in enhancing productivity, stress resistance, toxin reduction, and the development of specific traits within microbial strains. Moreover, it plays a crucial role in optimizing microorganisms for the increased production of valuable resources such as enzymes, biofuels, pharmaceuticals, and biomolecules. Through metabolic engineering, site-directed mutagenesis also strengthens the resistance of strains to environmental stresses while modifying metabolic pathways. Whether redirecting these pathways, engineering proteins, or mitigating unwanted by-products, site-directed mutagenesis empowers researchers to custom-tailor microorganisms to meet specific biotechnological and medical objectives, underlining its paramount importance in advancing scientific endeavours.
References:
- Hutchison CA, Phillips S, Edgell MH, Gillam S, Jahnke P, Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978;253(18):6551-6560. http://www.ncbi.nlm.nih.gov/pubmed/681366.
- Yang H, Li J, Du G, Liu L. Microbial production and molecular engineering of industrial enzymes: challenges and strategies. InBiotechnology of microbial enzymes 2017 Jan 1 (pp. 151-165). Academic Press..
- Carter P. Site-directed mutagenesis. Biochemical Journal. 1986 Jul 7;237(1):1.
