Introduction:
Nucleic acids, which are found in all living species, are the foundation of inheritance. The genotype is the sequence of nucleic acids that encodes the genetic information, which gives rise to the features manifested by the organism, the phenotype. The gene is the fundamental unit of information that is coded. The storing and expression of genetic information is a key role of nucleic acids.
There are two types of nucleic acids: Deoxyribonucleic acid (DNA) and Ribonucleic acid (RNA).
Extraction of nucleic acid:
There are several methods for extracting nucleic acids from biological samples, including cells, tissues, and organisms.
There are both various manual protocol and commercial kits available that are specifically designed for nucleic acid extraction, which often use a combination of chemical and mechanical methods to purify the nucleic acids from the sample.
The primary goal of extracting nucleic acids from a biological sample is to obtain a pure, high-quality sample of DNA that may be used for downstream applications such as PCR, sequencing, cloning, genetic analysis or forensic applications.
Nucleic acid extraction is the first step in each amplification experiment, irrespective of the type of amplification employed to identify a specific pathogen. It is a critical preanalytical stage in the development and performance of any effective molecular diagnostic technique.
Three major processes included in nuclide acid extraction:
- Isolation
- Purification
- Concentration
Determining DNA Purity and Yield:
Sample collection and preparation, specimen transport and storage, nucleic acid stability in samples, and nucleic acid extraction are all preanalytical testing variables.
Physical Examination
One can analyse or assess DNA purity and yield by observing the elutes or pellets over time after gaining a ton of experience in DNA extraction.
It is possible that the DNA has good quality and amount if it precipitates (appears as whitish pellets that are easily visible), resembling a cotton thread. Before continuing, though, a quality assessment using an instrumental analysis must be required.
UV absorbance (Optical density)/Spectrophotometry
Absorbance is the most commonly used approach for determining both nucleic acid concentration and purity. Absorbance measurements can be used to assess the quantity of DNA or RNA in purified materials. The nucleic acid sample is inserted in a quartz cuvette, which is subsequently placed inside the UV spectrophotometer. Nucleic acid concentrations can be determined directly by measuring absorbance values at 260 nm against blank using the principle of Beer Lambert. UV light that is transmitted through the sample at a specified path length.
The ratio of absorbance at 260 nm to 280 nm (A260/A280) is commonly used to assess the purity of the DNA. A ratio of around 1.8 indicates pure DNA, while ratios of less than 1.8 may indicate the presence of contaminants such as proteins or salts.
The NanoDropTM instrument, the DeNovix DS-11 spectrophotometer, and the Blue-Ray Bio EzDrop spectrophotometer are a few examples of small volume spectrophotometers that can analyze samples as little as 1 ul
Fluorometry
Fluorometry is similar to spectrophotometry but it uses a fluorescent dye that binds specifically to double-stranded DNA. Fluorometry is more sensitive than spectrophotometry and can detect small amounts of DNA. These techniques make use of fluorescent dyes that intercalate and bind nucleic acid grooves, bind DNA or RNA without specificity, or bind nucleic material with specificity. Fluorometry is also more specific and can detect the presence of other contaminants such as RNA and proteins.
Based on their spectral characteristics and mechanism of binding, several dyes are customized to measure dsDNA, ssDNA, RNA, or short RNAs (such as miRNA). The Qubit fluorometer is an example of molecular instrument developed and marketed by Invitrogen that is used for a measuring the concentration of nucleic acid which is based on the principle of fluorometry.
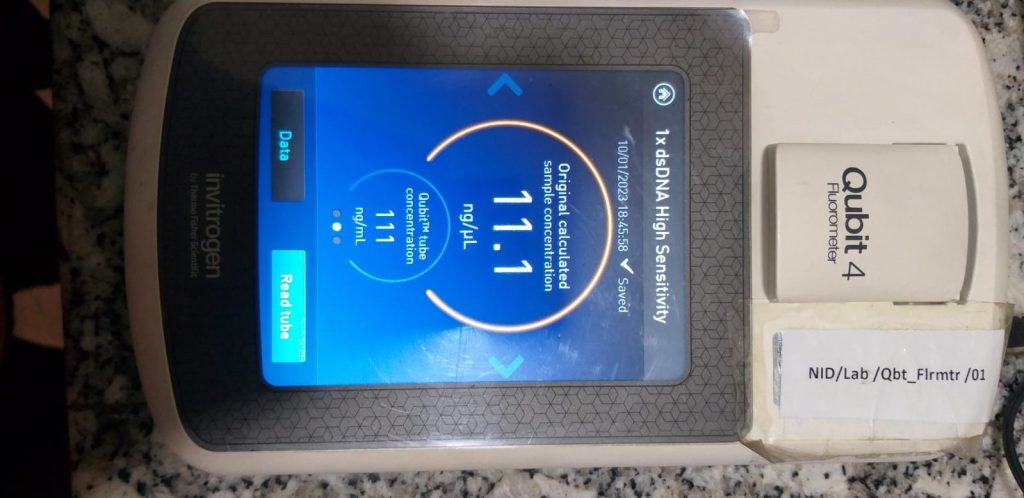
Fig: The observed reading in a Qubit fluorometer
Agarose gel Electrophoresis
Gel electrophoresis can be used for both qualitative and quantitative analysis of DNA. For qualitative analysis, a gel is stained with ethidium bromide or other DNA-specific dyes and visualized under UV light. This allows researchers to see the integrity of the DNA sample and to identify any degraded or fragmented DNA.
For quantitative analysis, researchers can use Gel electrophoresis by comparing the intensity of bands on the gel to a known amount of a DNA/gene/molecular ladder. The ladder is a mixture of DNA fragments of known sizes that are run on the gel alongside the sample. By comparing the intensity of the band on the gel to the band of the ladder, one can estimate the amount of DNA in their sample. This method is called gel-based quantification.
However, gel-based quantification is not very accurate or precise and it does not provide an exact concentration of DNA in the sample. Additionally, it depends on the quality of the gel, the staining and the visualization conditions.

Fig: The DNA band under the UV transilluminator
REAL-TIME PCR (qPCR)
Real-time PCR, also known as quantitative PCR (qPCR), uses fluorescent dyes that bind double-stranded DNA or fluorometric probes that bind target DNA during the annealing phase of PCR. The fluorescence signal is monitored as it amplifies using specialized heat cyclers with fluorescence detecting modules. The amount of DNA present is proportional to the measured fluorescence. The amounts of target DNA in test samples are calculated using a standard curve. Digital PCR techniques can also be used to calculate DNA amounts without the use of a standard curve.
In the case of RNA quantification, complementary DNA (cDNA), which is synthesized by reverse transcription of the RNA that serves as the PCR template.
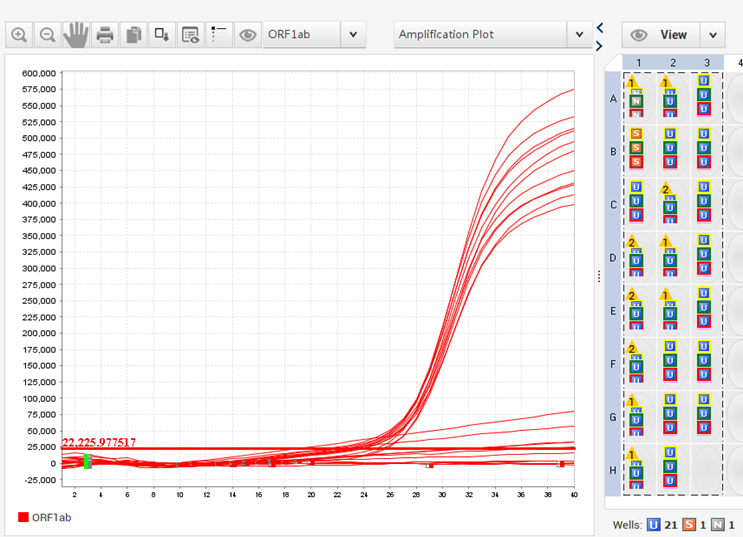
Fig: The curve observed in qPCR
Magnetic nanoparticles in nucleic acid detection
The term “nanoparticles” (NPs) refers to particles with a size between 1 and 100 nm that respond to being exposed to a magnetic field. Magnetic nanoparticles (MNPs) combine the features of both magnetic particles and NPs. MNPs are appropriate for nucleic acid detection in high-throughput samples requiring quick turnaround time due to their high binding capacity, specificity of binding, and fast magnetic response.
Due to limited time, it has also been challenging for traditional PCR-based detection technologies to keep up with the rising high-throughput demand in recent years for applications such as clinical in vitro diagnosis, mobile medicine, and emergency treatment of catastrophic infectious diseases.
MNPs can conduct magnetically regulated aggregation and dispersion, have a high surface area to volume ratio, and have a high binding rate with detecting compounds, which makes preconcentration, purification, and separation of nucleic acids simple and easy. Because of their excellent dispersibility, MNPs can bind biomolecules efficiently and fast.
Factors affecting the purity of nucleic acid:
Extraction method: Different extraction procedures can result in varying degrees of purity for the isolated nucleic acids. Although phenol-chloroform extraction and salting out are standard procedures for extracting nucleic acids, additional methods such as silica-based purification or column-based purification can also be utilized.
Type of sample: The type of sample also influences the purity of extracted nucleic acids. RNA extraction, for example, is often more difficult than DNA extraction because RNA is more labile and degrades more easily.
Sample quality: The purity of the extracted nucleic acids can be considerably influenced by the quality of the starting material. If the sample is contaminated with bacteria or other microbes, for example, it can result in high amounts of nucleases, which can destroy the nucleic acids. Likewise, subjecting the sample to high temperatures or other extreme circumstances might destroy the nucleic acids and impair their purity.
Quantity of starting material: The amount of starting material can also affect the purity of the extracted nucleic acids. If the sample is too small, the yield of nucleic acids may be low, which can lead to a lower purity.
Quality of reagents: The quality of the reagents used in the extraction process can also affect the purity of the nucleic acids. For example, if the reagents are expired or contaminated, it can lead to lower purity of the extracted nucleic acids.
Contamination: If the laboratory equipment or the work environment is not clean, it can lead to contamination of the nucleic acids.
The experience and expertise of the researcher: An expert researcher will be aware of potential sources of contamination and will take appropriate efforts to reduce them. They will also understand how to manage and store the isolated nucleic acids in order to keep them pure and of high quality. They will also have troubleshot experience for problems that may emerge during the extraction process, as well as the ability to recognize and solve issues that may influence the purity of the extracted nucleic acids.
Validation and calibration of the instruments used:
Validation is the process of verifying that the instruments employed in the extraction process are capable of performing the desired purpose. Instrument validation and calibration help to guarantee that the instruments are working properly and that the data obtained are accurate and dependable.
Applications:
- Forensic science
- Medical diagnostics
- Paternity Tests
- Plant and animal breeding
- Environmental monitoring
- Archaeology
- Biotechnology
- Hormones
- Vaccines
- Gene Therapy
- Sequencing
References:
- Shin JH. Nucleic Acid Extraction Techniques. Advanced Techniques in Diagnostic Microbiology. 2012 Apr 5:209–25.
- Killeen AA. Quantification of nucleic acids. Clin Lab Med. 1997 Mar;17(1):1-19.
- Barbas, C.F., Burton, D.R., Scott, J.K. and Silverman, G.J., 2007. Quantitation of DNA and RNA. Cold Spring Harbor Protocols, 2007(11), pp.pdb-ip47.
- https://worldwide.promega.com/
