Introduction:
- Enzymes are biological catalysts (also known as biocatalysts) that enhance biochemical reactions in living organisms that can be isolated from cells and used to catalyze a multitude of commercially relevant processes.
- Enzymes are the active proteins (except RNAse) that can catalyze biochemical reactions.
- Enzyme, a substance that works as a catalyst in living organisms, regulating the speed at which chemical reactions occur while maintaining unaltered.
- All biological activities in living things involve chemical reactions, and enzymes regulate the majority of them. Many of these processes would not occur at a significant rate without enzymes.
- Enzymes catalyze all aspects of cell metabolism.
- Enzymes are highly specialized catalysts with extraordinary catalytic power and also with remarkable specificity, catalyzing almost all cellular reactions. Therefore, they are known as the basis of life.
History:
The word “enzyme” is derived from the Greek words en (meaning “inside”) and zume (meaning “yeast”) and was first used by the German biologist Wilhelm Kühne in 1878 when he was characterizing the capacity of yeast to make alcohol from carbohydrates.
Nomenclature:
Enzyme nomenclature is drawn from their substrates or the catalyzed chemical reactions, and “ase” is frequently appended as a suffix.
Nucleases are enzymes that operate on nucleic acid. Dipeptidase is an enzyme that hydrolyzes dipeptides.
Exceptions: Trypsin, pepsin, and chymotrypsin
Few enzymes exist in their inactive forms, which are known as proenzymes or zymogens; for example, pepsin has pepsinogen as its zymogen.
According to the International Union of Biochemistry and Molecular Biology, enzymes can be indexed using letters and numbers: the letter EC plus four digits indicating four components. The first number comprises enzymes that are categorized based on the mechanism of enzymatic reaction.
Classification:
Broadly classified into Several categories
On the basis of location
1. Intracellular enzymes
Functional within the cells where they are synthesized.
Examples: LDH, AST, ALT, ALP, CK etc.
2. Extracellular enzymes
Active outside the cells.
Examples: All the digestive enzymes, e.g., amylase, lipase, maltase, sucrose, pepsin etc.
On the basis of structure
They are divided into two general categories:
Simple enzyme
It consists entirely of amino acids
Conjugated enzyme
It contains a non-protein group called a cofactor, which is required for biological activity.
When a cofactor is removed from a conjugated enzyme, a simple enzyme known as an apoenzyme is formed, which is usually physiologically inactive. Holoenzyme refers to a complete, biologically active conjugated enzyme (simple enzyme plus cofactor).
A cofactor can be covalently or non-covalently attached to the enzyme’s protein component. Some cofactors are simple metal ions, whereas others are complex organic groups, which are also known as coenzymes. Cofactors which are tightly associated with the protein covalently or non-covalently are called prosthetic group.
Classification of Enzymes by IUB System:
The International Union of Biochemistry (IUB) implemented policies for the nomenclature and classification of enzymes in 1964. IUB is now IUBMB (International Union of Biochemistry and Molecular Biology).
Each enzyme in the IUB system has a name and a special identification number.
According to the general rule, enzymes are primarily categorized and named based on the reaction they catalyze.
The unique attribute that separates one enzyme from another is the chemical reaction that specific enzyme catalyzed.
This unique characteristic of an enzyme is employed as the basis for enzyme classification and nomenclature.
Features
- Enzymes have been categorized into seven major groups.
- Each class is further subdivided into sub-subclasses and sub-subclasses.
- Enzymes are assigned a four-digit unique number called an EC number in this categorization.
- Enzyme Commission number, which serves as an identification code which is made up of four digits separated by periods.
Each digit indicates a specific category in the classification.
First digit – Major class.
Second digit – Sub-class.
Third digit – Sub-subclass.
Fourth digit – Systemic specific name of the enzyme.
E.g., Hexokinase (2.7.1.1)
Based on their action they are involved in; the enzymes are divided into 7 major classes. They are as follows:
1) Oxido-Reductases
This class of enzymes are involved in oxidation-reduction processes.
Here, oxygen and hydrogen atoms or electrons are transferred from one substrate to another; these substrates are known as oxidases.
E.g., Alcohol dehydrogenase.
2) Transferases
Transferases are enzymes that catalyze the transfer of functional groups such as amine, carboxyl, carbonyl, methyl, acyl, glycosyl, and phosphoryl.
E.g., Phosphorylases, carbamoyl transferase groups.
3) Hydrolases
These are enzymes that cause various compounds to hydrolyze.
Esterases or digesting enzymes, are hydrolases.
Esterases, such as phosphatases, that act on ester bonds, and proteases or peptidases that act on amide bonds in peptides are examples of hydrolases.
E.g., Lipase.
4) Lyases
Lyases are the enzymes that catalyze addition and elimination processes. The bond between a carbon atom and another atom, such as oxygen, sulfur, or another carbon atom, is broken by lyase-catalyzed processes.
E.g., Aldolase, dehydratase.
5) Isomerases
Enzymes are engaged in all isomerization reactions, or changes in the molecular form of a substrate.
E.g., Phosphotriose isomerase, bisphosphoglycerate mutase.
6) Ligases (Synthetases)
Enzymes that catalyze synthetic reactions involving the joining of two molecules and the consumption of ATP.
E.g., Succinate thiokinase, citric acid synthetase.
7) Translocases
These enzymes catalyze the passage of ions or molecules across membranes or their separation within membranes.
E.g., Ornithine translocase, carnitine-acylcarnitine translocase.
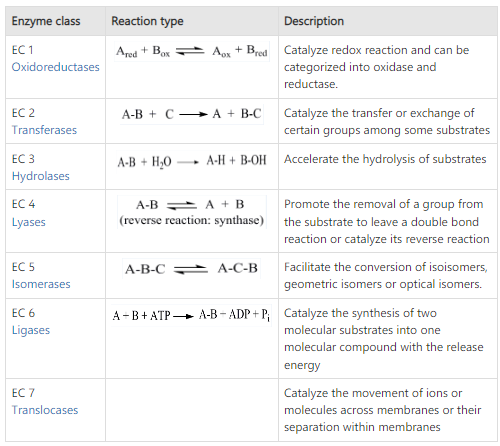
Fig: Classification of enzyme
Enzyme structure and substrate binding:
Active sites:
- The active site of an enzyme is the region where substrate molecules interact and undergo a chemical reaction.
- The active site is made up of amino acid residues that establish temporary bonds with the substrate (binding site) and residues that catalyze the substrate’s reaction (catalytic site).
- Although the active site accounts for only 10-20% of an enzyme’s volume, it is the most significant component because it directly catalyzes the chemical process.
- It is typically composed of three to four amino acids, with additional amino acids necessary within the protein to maintain the tertiary structure of the enzymes.
- Each active site evolves to be optimized to bind a single substrate and catalyze a specific reaction, resulting in great specificity.
- This specificity is determined by the arrangement of amino acids within the active site as well as the structure of the substrates. Enzymes must sometimes bind with cofactors in order to function effectively.
- The active site is typically a groove or pocket in the enzyme that can be found in a deep tunnel within the enzyme or between the interfaces of multimeric enzymes.
- The active site is a location on an enzyme to which the substrates of a chemical reaction bind in order to undergo a catalyzed chemical reaction. This region is divided into two sub-regions: a binding site and a catalytic site.
- An active site contains a binding site that binds the substrate and orients it for catalysis. The direction of the substrate and its close proximity to the active site are so crucial that the enzyme can still work properly even when all other portions are altered and lose function.
- The binding site contains certain residues that can aid in the binding of the substrate (reactants) to the enzyme.
- The catalytic site aids in catalyzing the chemical process. Furthermore, this area is quite little in comparison to the whole volume of the enzyme; around 10-20% of the total volume of the enzyme.
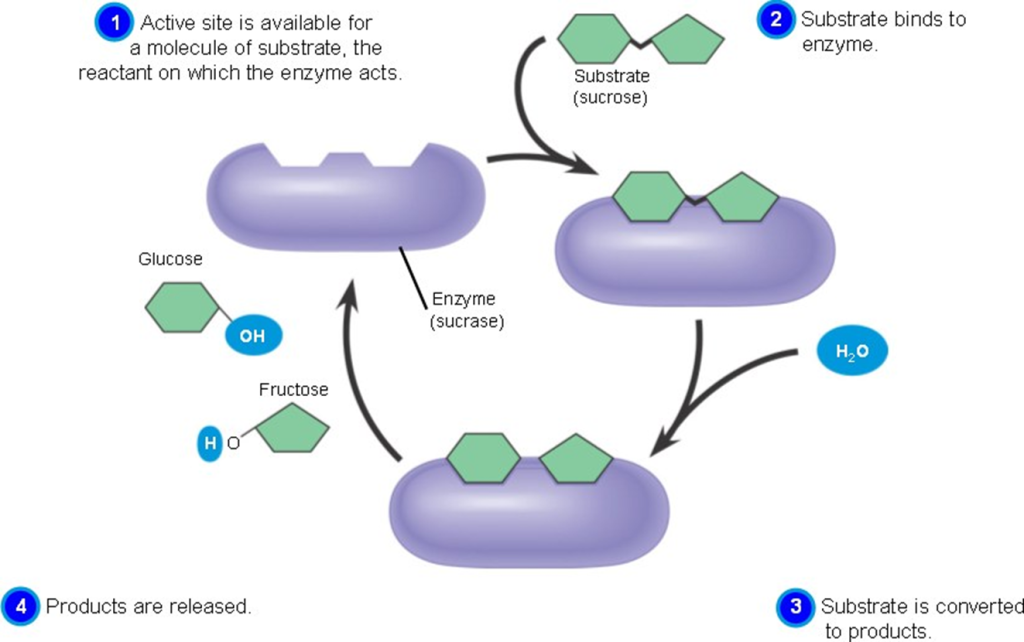
Fig: Active site and substrate binding
Initially, the contact between the active site and the substrate is non-covalent and transitory.
There are four major forms of interactions that retain the substrate in a fixed orientation and form an enzyme-substrate complex (ES complex):
Hydrogen bonds,
van der Waals interactions,
hydrophobic interactions and
electrostatic force interactions
The mechanism of enzymatic action:
Enzymes draw substrates to their active site, catalyze the chemical reaction that produces the products, and then eventually allow the products to separate (separate from the enzyme surface).
The enzyme-substrate complex is the combination formed by an enzyme and its substrates.
Electrostatic and hydrophobic forces draw substrates to the active site. They are called noncovalent bonds because they are physical attractions rather than chemical bonds.
When two substrates and one enzyme are involved, the complex is referred to as a ternary complex; when one substrate and one enzyme are involved, the complex is referred to as a binary complex.
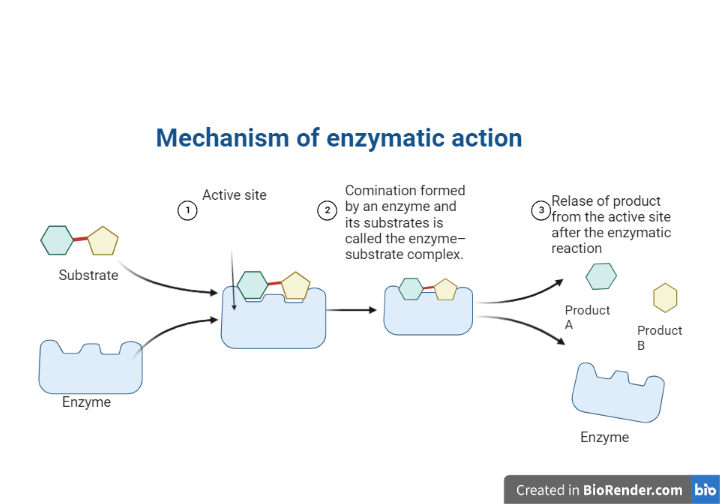
Fig: Mechanism of enzymatic reaction
Enzyme Specificity:
Each type of enzyme will typically act on only one substrate to catalyze a single biological reaction. Because different enzymes have different shaped active sites, they are specialized. The form of an enzyme’s active site is complementary to the shape of its specific substrate. This suggests they are the right shapes for each other.

Fig: Enzyme specificity
Absolute Specificity
An enzyme that catalyzes only one substrate reaction.
Group Specificity
An enzyme that catalyzes processes involving similar molecule containing the same functional group.
e.g., hexokinase
Linkage Specificity
An enzyme that catalyzes the formation of breakage of only certain binds in molecule.
e.g., proteases – hydrolyzes peptide bonds
Stereochemical Specificity
An enzyme capable of differentiating between two enantiomers.
Hexokinases always act on D-glucose not L-glucose.
Applications:
Medicine: Enzymes are used in the production of drugs and as therapeutic agents to treat various diseases. For example, enzymes are used to break down clots in the blood to prevent heart attacks and strokes, and to digest food in people with digestive disorders.
Industry: Enzymes are used in the production of various consumer products, such as detergents, textiles, and biofuels. They are also used in the food industry to produce cheese, bread, and other products.
Research: Enzymes are widely used in research to study biological processes, including DNA replication, protein synthesis, and metabolism. They are also used in diagnostic tests to detect the presence of certain molecules or conditions in samples.
Environmental: Enzymes are used in the treatment of waste and in the production of biofuels, which can help reduce the reliance on fossil fuels and decrease the carbon footprint of various industries.
Agriculture: Enzymes are used in the production of animal feed and in the processing of crops to improve efficiency and yield.
Energy: Enzymes are being explored as a potential source of renewable energy, as they can convert organic matter into biofuels.
References:
- Harrow, B., and Mazur, A.: Textbook of Biochemistry, 109, Saunders, Philadelphia (1958).
- Pfeiffer, J.: Enzymes, the Physics and Chemistry of Life, pg 171-173, Simon and Schuster,NY (1954)
- Hedstrom, L., 2001. Enzyme specificity and selectivity. e LS.
- Bugg, T.D., 2012. Introduction to enzyme and coenzyme chemistry. John Wiley & Sons.
- Boyce, S. and Tipton, K.F., 2001. Enzyme classification and nomenclature. e LS.
- Buchholz, K., Kasche, V. and Bornscheuer, U.T., 2012. Biocatalysts and enzyme technology. John Wiley & Sons.
- McDonald, A.G., Boyce, S. and Tipton, K.F., 2015. Enzyme classification and nomenclature. eLS, pp.1-11.
