Introduction:
- DNA microarray technology (also known as DNA arrays, DNA chips, or biochips) is one of the most recent and significant advances in experimental molecular biology.
- Microarrays are laboratory instruments used to detect the expression of hundreds of genes at once and to identify the genes present in the genome of an organism. It uses nucleic acid hybridization properties to monitor DNA or RNA abundance on a genomic scale in many types of cells.
- Microarrays are extremely useful to scientists because they allow them to study a huge number of genes in a short amount of time.
- Microarrays are used in medical diagnosis and therapy (e.g., comparing healthy cells to diseased cells to determine disease features), biotechnology (e.g., in agriculture to identify insect pests), and criminal investigation (forensic analysis)
- The microarray is made up of a small (typically 2cm2) piece of glass, plastic, or silicon (also known as chips) with probes linked to a location (called a gene spot) in a grid pattern. There may be 10,000 or more spots per cm2
- Probes are short pieces of single-stranded DNA (oligonucleotides) or RNA that are designed to be complementary for a given base sequence (this sequence depends on the purpose of the microarray).
What is DNA microarray technology?
This method is used to do parallel gene expression analysis on hundreds of genes with known and unknown functions, as well as DNA homology analysis to find polymorphisms and mutations in both prokaryotic and eukaryotic genomic DNA.
However, a DNA microarray is an orderly arrangement of thousands of identifiable sequenced genes printed on an impermeable solid support, typically glass, silicon chips, or a nylon membrane.

Fig: Microarray
Each recognized sequenced gene on the glass, silicon chips, or nylon membrane belongs to a single gene and corresponds to a fragment of genomic DNA, cDNAs, PCR products, or chemically produced oligonucleotides of up to 70mers.
A single DNA microarray slide/chip may typically include thousands of spots, each representing a single gene and together representing an organism’s whole genome.
Principle:
The DNA microarray technology principle is predicated on the concept that complementary DNA sequences can be employed to hybridize immobilized DNA molecules.
This involves three major multi-stage steps:
Manufacturing of microarrays:
This step involves the availability of a chip or a glass slide with its special surface chemistry, the robotics used for producing microarrays by spotting the DNA (targets) onto the chip or for their in-situ synthesis.
Sample preparation and array hybridization step:
This step involves mRNA or DNA isolation followed by fluorescent labelling of cDNA probes and hybridization of the sample to the immobilized target DNA.
Image acquisition and data analysis:
Finally, this step involves microarray scanning, and image analysis using sophisticated software programs that allows us to quantify and interpret the data. However, here, we will concentrate on how microarray experiments are performed in the laboratory, rather than the technological developments involving array construction or manufacturing using precision robotic devices.
Steps:
Sample preparation and probe labelling
A DNA microarray sample can be processed and labeled in a variety of ways. The sample preparation process begins with the isolation of a total RNA containing messenger RNA, which ideally reflects a quantitative copy of genes expressed at the time of sample collection. This step is critical since the overall performance of any microarray experiment is dependent on the quality of the RNA.
The microarray sample being evaluated, whether it is mRNA for a gene expression study or DNA generated from genomic analysis, is transformed to a labelled population of nucleic acids, the probe.
These probes are usually made up of thousands of distinct tagged nucleic acid fragments. Fluorescent dyes, particularly the cyanine dyes Cy3 and Cy5, have been used as the primary label in microarray analysis. Fluorescence has the advantage of allowing the detection of two or more distinct signals in a single experiment.
Hybridization
Hybridization is the process of joining two complementary strands of DNA to form a double-strande molecule. Here, the labelled cDNA (Sample and Control) is mixed together, and then purified to remove contaminants such as primers, unincorporated nucleotides, cellular proteins, lipids, and carbohydrates.
Two or more samples labeled with distinct fluorescent dyes can be hybridized at the same time, resulting in simultaneous hybridization at each target site. The relative abundance of various sequences in each of the samples can be calculated by analyzing the varied fluorescence signals associated with each place.
Washing
Before hybridization however, the microarray slides are incubated at high temperature with solutions of saline-sodium buffer (SSC), Sodium Dodecyl Sulfate (SDS) and bovine serum albumin (BSA) to reduce background due to nonspecific binding. The slides are washed after hybridization, first to remove any labelled cDNA that did not hybridize on the array, and secondly to increase stringency of the experiment to reduce cross hybridization. The latter is achieved by either increasing the temperature or lowering the ionic strength of the buffers.
Image acquisition (Scanning) and Data analysis
Microarray scanners typically comprise two separate lasers that generate light at wavelengths suitable for stimulating the fluorescent dyes used as labels. A confocal microscope attached to a detector system records the emitted light from each of the microarray spots, allowing high-resolution detection of the hybridization signals. Despite their small size, microarrays provide a huge amount of data from a single experiment.

Fig: Microarray experimental principles
Types:
There are now two commercially available DNA microarray platforms/types.
- Glass DNA microarrays which involve the micro spotting of pre-fabricated cDNA fragments on a glass slide.
- High-density oligonucleotide microarrays often referred to as a “chip” which involves in situ oligonucleotide synthesis.
cDNA-based microarrays
Glass DNA microarrays was the first type of DNA microarray technology developed.
These microarrays use c-DNA probes with lengths between 500 and 1000 bp.
oligose probes with lengths between 25 and 100 mers, or fragments of PCR products.
Two samples are merged on a single slide in this approach to provide a relative expression level for each area.
Oligonucleotide based microarrays
They typically have a probe length of 18 to 30 mers.
Shorter probe lengths allow less errors during probe synthesis.
oligonucleotide probes need to be carefully designed so that all probes acquire similar melting temperatures (within 50 c) and eliminate palindromic sequences. In situ (on chip) oligonucleotide array format is a sophisticated platform of microarray technology which is manufactured by using the technology of in situ chemical synthesis that was first developed
by Stephen Fodor.
However, Affymetrix, the industry leader in in situ oligonucleotide microarrays, has further pioneered this type of technology to make so-called GeneChips, which are high-density oligonucleotide-based DNA arrays.
Affymetrix’s GeneChips are manufactured using photolithography and combinatorial chemistry to create small single strands of DNA on 5-inch square quartz wafers.
Applications:
- To compare which genes are being expressed using microarrays the following steps occur:
- mRNA is collected from both types of cells and reverse transcriptase is used to convert mRNA into cDNA
- PCR may be used to increase the quantity of cDNA (this occurs for all samples to remain proportional so a comparison can be made when analysis occurs)
- Fluorescent tags are added to the cDNA
- The cDNA is then denatured to produce single-stranded DNA
- The single-stranded DNA molecules are allowed to hybridize with the probes on the microarray
- When the ultraviolet light is shone on the microarray the spots that fluoresce indicate that gene was transcribed (expressed) and the intensity of the light emitting from the spots indicates the quantity of mRNA produced (i.e., how active the gene is). If the light being emitted is of high intensity, then many mRNA were present, while a low intensity emission indicates few mRNA are present
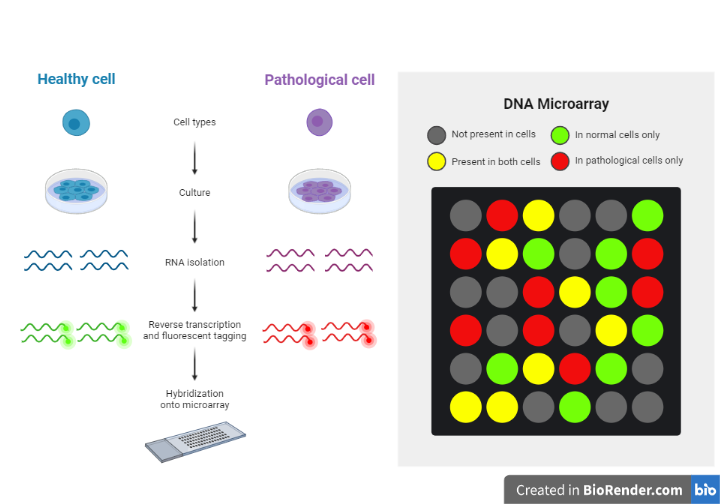
Fig: Applications of microarray analysis
- Identifying protein binding sites;
- Determining sub-cellular localization of gene products;
- Cancer research: Molecular characterization of tumors on a genomic Scale, more reliable diagnosis and effective treatment of cancer.
- Immunology: Study of host genomic responses to bacterial infections.
References:
- Bowtell, D., Sambrook, J. and Manual, A.D.C., 2003. DNA microarrays. Cold Spring Habor Laboratory Press, New York, 79.
- Bumgarner, R., 2013. Overview of DNA microarrays: types, applications, and their future. Current protocols in molecular biology, 101(1), pp.22-1.
- Sanjeev Singh, Megha Kadam Bedekar (2014). DNA Microarray and its Applications in Disease Diagnosis. Volume 3 Issue 12, December 2014
- Stoughton, R.B., 2005. Applications of DNA microarrays in biology. Annu. Rev. Biochem., 74, pp.53-82.
- Schena, M. ed., 1999. DNA microarrays: a practical approach (No. 205). Practical approach series.
- https://www.thermofisher.com/
