Introduction:
- DNA barcoding is a method of identifying species using a short, standardized DNA sequence from a specific region of an organism’s genome. The technique involves analyzing the DNA of an unknown specimen and comparing it to a reference database of known DNA sequences to determine the identity of the species.
- DNA barcoding has a number of applications in the fields of biology, conservation, and environmental science. For example, it can be used to identify species in a mixed sample, such as a group of insects collected from a specific location. It can also be used to confirm the identity of a species in cases where traditional identification methods are difficult or impossible, such as when dealing with small or damaged specimens.
- DNA barcoding is a relatively fast and inexpensive method of species identification, and it has the potential to revolutionize the way we study and monitor biodiversity. However, there are also limitations to the technique, including the need for a reference database of known DNA sequences and the possibility of misidentification due to the presence of DNA from multiple species in a sample.
History:
The idea of using DNA to identify species dates back to the late 1960s, when researchers first proposed using DNA sequences as a way to identify species. However, it wasn’t until the early 2000s that DNA barcoding became widely recognized as a useful tool for species identification.
In 2003, Paul Hebert, a professor at the University of Guelph in Canada, published a paper outlining the concept of DNA barcoding, in which he proposed using a standardized DNA region called the “barcode region” to identify species. The barcode region is typically a small section of the genome that is highly variable among different species, but relatively conserved within a species. The most commonly used barcode region is the cytochrome c oxidase subunit 1 (CO1) gene, which is found in the mitochondria of most organisms.
Since the publication of Hebert’s paper, DNA barcoding has become widely used in the fields of biology, conservation, and environmental science. It has been used to identify species in a variety of contexts, including in mixed samples, in small or damaged specimens, and in environmental samples. DNA barcoding has also been used to monitor changes in biodiversity over time and to assess the impact of human activities on ecosystems.
Types:
There are several types of DNA barcoding that are used for different purposes.
Standard DNA barcoding: This is the most common type of DNA barcoding and involves analysing a standardized DNA region called the “barcode region” to identify species. The barcode region is typically a small section of the genome that is highly variable among different species, but relatively conserved within a species. The most commonly used barcode region is the cytochrome c oxidase subunit 1 (CO1) gene, which is found in the mitochondria of most organisms.
Multilocus DNA barcoding: This approach involves analyzing multiple DNA regions to identify species. The advantage of this method is that it can provide more accurate identifications, especially in cases where the barcode region is not sufficiently variable to distinguish between species.
Environmental DNA (eDNA) barcoding: This type of DNA barcoding involves analyzing DNA from environmental samples, such as water or soil, to identify the species present in an ecosystem. eDNA barcoding can be used to detect the presence of rare or elusive species and to monitor changes in biodiversity over time.
DNA metabarcoding: This approach involves analyzing DNA from mixed samples, such as the contents of an animal’s gut, to identify all of the species present in the sample. DNA metabarcoding can be used to study the diet and feeding habits of animals, as well as the biodiversity of ecosystems.
Methods:
Polymerase chain reaction (PCR): This method involves amplifying the DNA region of interest using specialized enzymes. PCR allows researchers to obtain enough DNA for analysis even if the starting sample is small or degraded.
DNA sequencing: Once the DNA region of interest has been amplified using PCR, it can be sequenced to determine the exact order of the DNA base pairs. DNA sequencing can be performed using a variety of techniques, including Sanger sequencing and next-generation sequencing.
DNA fragment analysis: This method involves separating DNA fragments based on size using techniques such as electrophoresis or capillary electrophoresis. The resulting DNA fragment profile can be used to identify species or to determine the genetic relationships between organisms.
Bioinformatics: Once the DNA sequence has been obtained, it can be analyzed using computer algorithms to compare it to known sequences in a reference database. This allows researchers to determine the identity of the species based on the closest matching sequence.
Steps:
It is important to note that the specific protocol may vary depending on the specific DNA barcoding method being used and the goals of the study. The general protocol for DNA barcoding typically encompasses the following steps:
Sample collection: The first step in DNA barcoding is to collect a sample from the organism of interest. This may involve collecting a tissue sample, such as a leaf or a piece of fin tissue, or collecting a whole specimen, such as an insect or a fish.
DNA extraction: The next step is to extract the DNA from the sample. This can be done using a variety of techniques, such as phenol-chloroform extraction or automated DNA extractors.
PCR amplification: Once the DNA has been extracted, it is typically amplified using the polymerase chain reaction (PCR) to obtain enough DNA for analysis. The PCR reaction is typically performed using specific primers that are designed to amplify the DNA region of interest.
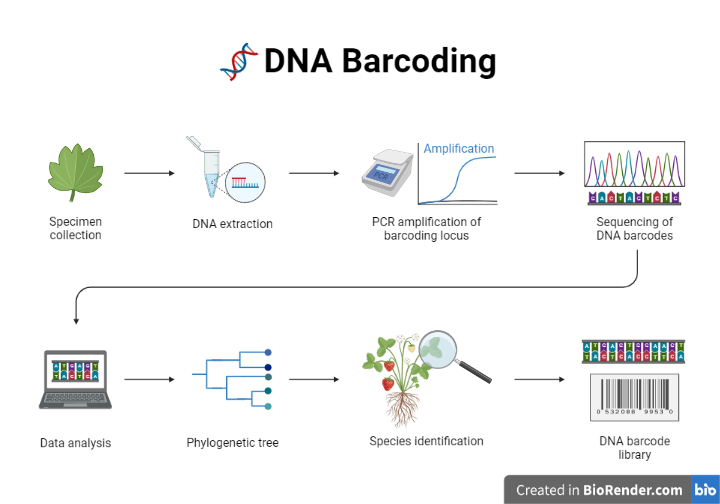
Fig: Steps of DNA Barcoding
DNA sequencing: The amplified DNA is then sequenced to determine the exact order of the DNA base pairs. This can be done using a variety of techniques, including Sanger sequencing and next-generation sequencing.
Data analysis: Once the DNA sequence has been obtained, it is compared to a reference database of known DNA sequences to determine the identity of the species. This can be done using computer algorithms that compare the DNA sequence to known sequences and determine the closest matching species.
Applications:
DNA barcoding has a number of applications in the fields of biology, conservation, and environmental science.
Species identification: DNA barcoding can be used to identify species in a mixed sample, such as a group of insects collected from a specific location. It can also be used to confirm the identity of a species in cases where traditional identification methods are difficult or impossible, such as when dealing with small or damaged specimens.
Biodiversity monitoring: DNA barcoding can be used to monitor changes in biodiversity over time, such as the presence or absence of particular species in an ecosystem.
Environmental impact assessment: DNA barcoding can be used to assess the impact of human activities on biodiversity, such as the effects of pollution or habitat destruction.
Food safety: DNA barcoding can be used to identify species in food products, such as fish and meat, to ensure that they are properly labeled and to detect fraudulent labeling.
Forensics: DNA barcoding can be used to identify species in forensic samples, such as animal hairs or tissue found at a crime scene.
Difference between DNA fingerprinting and DNA barcoding:
DNA fingerprinting and DNA barcoding are both powerful tools for identifying individuals and species, but they are used for different purposes and involve different DNA regions and reference databases. DNA fingerprinting and DNA barcoding are similar in that they both involve analyzing DNA to identify individuals or species.
Purpose: DNA fingerprinting is typically used to identify individuals, whereas DNA barcoding is used to identify species.
DNA region analyzed: DNA fingerprinting typically involves analyzing a large number of DNA regions to create a unique “fingerprint” for each individual. DNA barcoding, on the other hand, involves analyzing a standardized DNA region called the “barcode region” that is highly variable among different species, but relatively conserved within a species.
Reference database: DNA fingerprinting involves creating a reference database of known DNA fingerprints and comparing them to the DNA of an unknown individual to determine the identity of the individual. DNA barcoding involves comparing the DNA of an unknown specimen to a reference database of known DNA sequences to determine the identity of the species.
Applications: DNA fingerprinting is most commonly used in forensic science and in paternity testing, whereas DNA barcoding is used in a variety of fields, including biology, conservation, and environmental science.
References:
- Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003 Feb 7;270(1512):313-21
- Frézal, L. and Leblois, R., 2008. Four years of DNA barcoding: current advances and prospects. Infection, Genetics and Evolution, 8(5), pp.727-736.
- Moritz, C. and Cicero, C., 2004. DNA barcoding: promise and pitfalls. PLoS biology, 2(10), p.e354.
- Valentini, A., Pompanon, F. and Taberlet, P., 2009. DNA barcoding for ecologists. Trends in ecology & evolution, 24(2), pp.110-117.

