Introduction:
A genetic map is a schematic representation of the various genetic markers in the specific order in which they are located in a chromosome as well as the relative distances between these markers. Genetic mapping is the process of determining the physical position of a genetic component on chromosomes utilizing recombination and genetic markers.
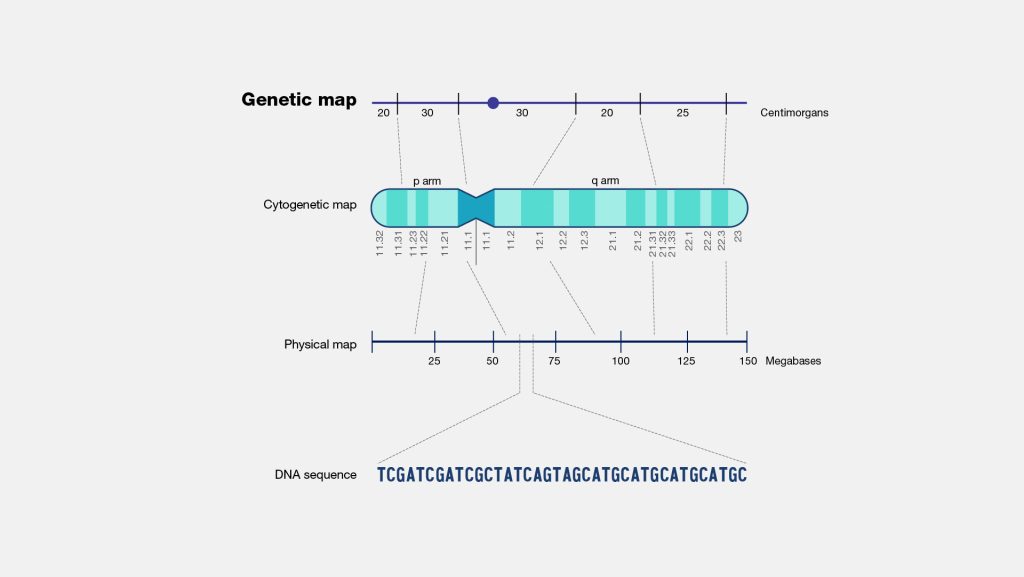
Fig: Genetic Map
Source: https://www.genome.gov/
Types:
Three different strategies have been used to construct genetic maps, which is usually referred to as linkage maps, cytogenetic or cytological maps and physical maps.
Linkage Maps
A linkage maps depicts the order of genetic markers and the relative distances between them as measured by terms of recombination frequencies between the markers. A map unit or centimorgan (cM) is that distance, which allows 1 % recombination between the concerned genes.
Cytogenetic Maps
A cytogenetic map depicts the location of various genes in a chromosome relative to specific microscopically visible landmarks in the chromosome. In most cases, each chromosome has a distinct banding pattern, which may be naturally present, as in Drosophila polytene chromosomes, or more commonly generated by specific staining protocols, as in the case of human chromosomes, where genes are mapped cytologically relative to these bands. Cytogenetic mapping is far more refined in organisms like Drosophila because their polytene chromosomes have a very detailed banding pattern. It is commonly used in eukaryotes since they have relatively large microscopically observable chromosomes.
Cytogenetic mapping is often used as a first step in localization of genes in plants and animals, which is then followed by physical mapping. In most species, cytogenetic mapping is only accurate within limits of about 5 mega base pairs (Mb=106 bp) along a chromosome; the resolution is much better in species that have polytene chromosomes.
For example, the sc gene of Drosophila is located at band 1B3 of X chromosome, while gene w is located in band 3c2. Cytogenetic mapping can be done using any one or more of several approached described below:
Fluorescence in situ hybridization (FISH)
In situ hybridization (ISH) is widely used to map the cytological locations of genes and other DNA sequences within large eukaryotic chromosomes. In the approach, the DNA of concerned gene/DNA sequence is used as probe. The probes are usually fluorescently labelled so that this form of in situ hybridization is called fluorescent in situ hybridization (FISH).
A fluorescent molecule absorbs light at a particular wavelength and that emits light at a longer wavelength. A fluorescent microscope having filters that transmit light within the emission wavelengths of the fluorescent molecules, is used to detect the light emitted by fluorescence. Since the probe binds only to the specific sequence of a chromosome, fluorescence generates a colorful glowing band on the chromosome against a colorless dark background; as a result, the slightest amount of colour is readily detectable. This makes FISH a very sensitive approach. FISH uses probes of 1 kb or greater size; usually, probe are 40kb or larger.
A unique advantage offered by FISH is as follows. Several (up to six) different DNA sequences can be simultaneously used in FISH to determine their relative locations in the concerned chromosomes. Each of these probes is labelled with a different combination of fluorescent molecules, so that each of them produces a distinctly colored and recognizable band on the chromosome. Computer imaging methods may be employed to detect and assign each fluorescently labelled probe a different colour. This approach is commonly called multicolor FISH (McFISH).
Somatic cell hybridization
It has been used to map genes on particular human chromosomes. In some cases, this method can map genes to more specific regions of human chromosomes,
For example, when some somatic hybrids contain human chromosomes that have suffered a large deletion that can be detected cytologically, genes located in the deleted segment can be confidently mapped this region. Similarly, when a human chromosome segment is translocated onto a rodent chromosome, the gene located in the translocated segment can be mapped.
Analysis of minor changes in polytene chromosomal structure
In this approach, small changes in polytene chromosome structure, e.g., small deletions are analyzed and correlated with the expression of specific genes. A comparison of findings from several strains differing slightly for the deleted regions often allows location of specific genes at specific chromosome bands.
Physical Maps
In a physical map, gene are depicted in the same order as they occur in chromosomes, and the distances between them are shown as number of base pair separating the genes. For example, genes sc and w of Drosophila are separated by about 1.5 X 106 bp.
In contrast, the distances between genes in a linkage map are shown as the frequency of recombination between them. Physical mapping usually requires the
- Cloning of many pieces of chromosomal DNA.
- Characterization of these fragments for size. i.e., length in base pairs, and the genes present in them.
- Determination of their relative location along a chromosome, (e.g., by Mc FISH). The ultimate of physical mapping is the complete sequence of a genome; this involves the following steps.
- Isolation of individual chromosomes using pulse-field gel electrophoresis (PFGE) or by fluorescence-activated chromosome sorting (FACS).
- Construction of chromosome specific libraries
- Identification of the complete contiguous series of clones that span the whole chromosome. Such a series of clones that contain overlapping (contiguous) pieces of chromosomal DNA constitutes a contig. Contigs can be created either by chromosome jumping and chromosome walking or by alignment of sequence-tagged sites (STSs) and yeast artificial chromosome (YAC) clones.
- Each YAC clone is subcloned, contig subclones are created, and ultimately the contig subclones are completely sequenced.
- The sequence of DNA fragments presents in contig subclones of a YAC clone are pieced together to yield the complete base sequence of the DNA fragment present in this YAC clone.
- Finally, the base sequences of all YAC clones constituting the contig for a chromosome are aligned to yield the complete base sequence of the chromosome.
