Introduction:
- A vaccine is a substance that trains the body’s immune system to recognize and protect against a disease caused by an infectious agent. Diseases causing agents are injected in the body in a weakened or dead form to give an easy target to body’s immunize system for practice.
- In conventional vaccinology, pathogenic organism is growth in the laboratory and from which a limited number of antigens are isolated. In contrast, reverse vaccinology is a latest computer-based approach for the discovery of antigen to design vaccines from genomic information with the help of bioinformatics. However, it can only identify only protein-based antigens that aims to identify putative vaccine candidates in the protein coding genome (proteome) of pathogens.
- Moreover, this technique has significantly reduced the time and also offers the possibility of using genomic information that are derived from computational/Insilco approach. Likewise, it is prominently useful for the potential antigens of pathogens which are difficult to culture.
History:
- Vaccine development has evolved over centuries, in 1796, Edward Jenner introduced the first successful smallpox vaccine and eventually, throughout the 20th century, several vaccines were developed for various diseases such as polio, measles, and influenza, significantly reducing their prevalence and impact.
- In this scenario, “reverse vaccinology” was proposed with a novel approach in vaccine development in 2000 by Rappuoli that represents a genome-based approach to vaccine development.
- A new approach to vaccine design was allowed after the access of the genomic information of microorganisms in 1995. Eventually, the vaccine against serogroup B meningococcus was the first reported successful application of reverse vaccinology. Since then, technique has been used in the development of vaccines against diseases such as group B streptococcus and Streptococcus pneumoniae.
Principle:
This computational approach is a branch of vaccinomics that is used to design vaccines predicting epitopes from the genetic information of the pathogens. The principle of reverse vaccinology involves the availability of the whole genome sequence of the organism and uses computational analysis to identify genes that encode proteins on the surface of the pathogen that are involved in the immune system. Likewise, this information is used to identify the potential antigens of vaccines Furthermore, antigenicity and immunogenicity of these potential antigens are tested to determine their suitability as a possible vaccine candidate. The technique is particularly useful for designing vaccines against highly infectious pathogens that are hazardous to culture in the laboratory.
Method:
Genomic study, prediction, and analysis
It initiates with the study and analysis of the entire lists of protein from the whole genomic information of the respective pathogen and uses computational approach to predict possible epitopes and other surface proteins responsible for immune system.
Computational approach
Several computational tools and algorithms are used for the prediction of epitope, testing the potential antigenic and immunogenic property, and comparing the genetic structures of pathogenic and non-pathogenic strains of the same species. These includes EpiMatrix, BlastiMer, ProPred, OptiMatrix, ClustiMer, Conservatrix, RANKPEP, and Vaccine CAD.
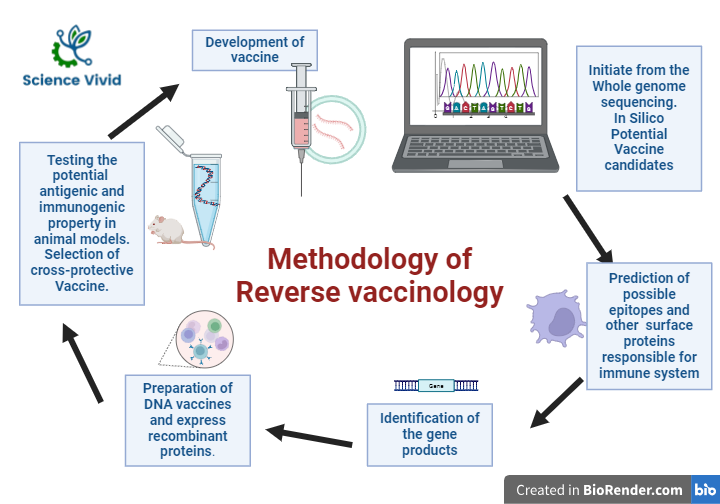
Fig: Methodology of Reverse Vaccinology
Laboratory examination and clinical trails
Furthermore, the selected antigens will be immunized in the animal model in order to examine if the antigens can induce an immune response. And if it evokes the expected immunological response, the selected antigens will be manufactured on a large scale for the clinical trails and other validation procedures and protocols.
Applications:
- Reverse vaccinology has been used to develop vaccines against bacterial pathogens, such as serogroup B meningococcus, group B streptococcus, and Streptococcus pneumoniae, malaria, Mycobacterium tuberculosis, syphilis, R. prowazekii, C. pneumoniae, N. meningitidis and Hepatitis C virus.
- By designing peptide pools to focus on specific HLA types or supertypes, reverse vaccinology allows for the identification of immunodominant protein antigens and the tackling of larger genomes that were previously difficult to study.
Traditional vaccinology vs. Reverse vaccinology:
| Conventional vaccinology | Reverse vaccinology | |
| Essential features | Most abundant antigens during disease | All antigens immunogenic during disease |
| Antigens immunogenic during disease | Antigens even if not immunogenic during disease | |
| Cultivable microorganism | Antigens even in non-cultivable microorganisms | |
| Animal models essential | Animal models essential | |
| Correlates of protection useful | Correlates of protection very important. Correct folding in recombinant expression important. High-throughput expression/analysis important. | |
| Advantages | Polysaccharides may be used as antigens | Fast access to virtually every single antigen |
| Lipopolysaccharide-based vaccines are possible | Non-cultivable microorganisms can be approached | |
| Glycolipids and other CD1-restricted antigens can be used | Non abundant antigens can be identified. Antigens that are not immunogenic during infection can be identified. Antigens that are transiently expressed during infection can be identified. Antigens not expressed in vitro can be identified. Non-structural proteins can be used. | |
| Disadvantages | Long time required for antigen identification. Antigenic variability of many of the identified antigens. Antigens not expressed in vitro cannot be identified. Only structural proteins are considered | Non proteic antigens cannot be used (polysaccharide, lipopolysaccharides,glycolipids and other CD1-restricted antigens) |
Source: Reverse vaccinology Rappuoli
