Introduction:
RNA splicing is the process by which cells remove introns (non-coding sequences) from a primary RNA transcript and join the remaining exons (coding sequences) together to create a mature RNA molecule. This process is important because it allows cells to generate multiple different proteins from a single gene by including or excluding different exons.
RNA splicing is an important post-transcriptional step required to create a mature mRNA, was first identified in the late 1970s. This process allows to generate a diversity of proteins from a limited number of genes.
RNA splicing occurs in both prokaryotes (bacteria and archaea) and eukaryotes (animals, plants, and fungi) and is an important process that allows cells to generate multiple different proteins from a single gene by including or excluding different exons. However, there are some differences between RNA splicing in prokaryotes and eukaryotes RNA splicing occurs in both prokaryotes and eukaryotes, there are some differences in the mechanisms and processes involved.

Fig: RNA splicing
Spliceosome: The spliceosome is the enzyme responsible for recognizing specific sequences at the ends of introns and removing them from the primary RNA transcript. In prokaryotes, the spliceosome is a simpler, ribonucleoprotein complex that consists of small RNA molecules called ribozymes and proteins. In eukaryotes, the spliceosome is a more complex structure that includes both small RNA molecules and larger proteins.
Intron removal mechanism: In eukaryotes, introns are removed by the spliceosome in a process called transesterification. In prokaryotes, introns are removed by a process called self-splicing, in which the intron itself acts as a ribozyme to cut itself out of the primary RNA transcript.
Alternative splicing: Alternative splicing is the process by which cells can produce multiple different proteins from a single gene by including or excluding different exons. This process is more common in eukaryotes than in prokaryotes.
Intron:
Introns are non-coding regions of DNA that are found within eukaryotic genes. They are present in the primary sequence of DNA, but are removed (or “spliced out”) during the process of gene expression to create the final mature RNA molecule. Introns are found in both protein-coding genes and non-coding RNA genes, and they play a variety of roles in the regulation of gene expression. Introns can be quite large, and they may contain regulatory sequences that control the expression of the gene in which they are found. Introns are an important part of the structure of eukaryotic genomes, and they contribute to the diversity and complexity of eukaryotic organisms.
- Introns do not contain information needed to encode proteins. They are removed from the RNA molecule before it is translated into a protein.
- Introns can vary significantly in size. Some are only a few dozen base pairs long, while others can be hundreds of thousands of base pairs long.
- Introns are typically found within genes, and they may be present in both protein-coding genes and non-coding RNA genes.
- Introns are removed from the primary RNA transcript through a process called splicing. This process involves the removal of the intron and the ligation (or joining) of the exons on either side of the intron.
- Introns can contain regulatory sequences that control the expression of the gene in which they are found. These sequences may influence the splicing of the RNA transcript or the stability of the RNA molecule.
- Introns can evolve more rapidly than exons, which may contribute to the diversity and complexity of eukaryotic genomes.
Exon:
Exons are the coding regions of DNA that are retained in the final RNA molecule and translated into protein. They are found within genes, and they contain the information needed to encode proteins. Exons are separated by introns in the primary RNA transcript, and during the process of gene expression, the introns are removed and the exons are joined (or ligated) to create the final, mature RNA molecule. Exons may be present in both protein-coding genes and non-coding RNA genes, and they play a crucial role in the synthesis of proteins, which are important for many functions in the cell and the organism as a whole.
- Exons contain the information needed to encode proteins. They are retained in the final RNA molecule and are translated into protein.
- Exons can vary in size, with some being only a few dozen base pairs long and others being several thousand base pairs long.
- Exons are typically found within genes, and they may be present in both protein-coding genes and non-coding RNA genes.
- Exons are separated by introns in the primary RNA transcript. During the process of splicing, the introns are removed and the exons are joined (or ligated) to create the final, mature RNA molecule.
- Exons tend to be more conserved (or less prone to change) than introns, which may reflect the important functional role that exons play in encoding proteins.
- Exons can evolve more slowly than introns, which may help to maintain the functional integrity of proteins.
Noncoding RNAS:
RNA molecules called non-coding RNAs (ncRNAs) are those that do not code for proteins. They are involved in a variety of cellular processes, including the regulation of gene expression, the modification of chromatin structure, and the processing of other RNA molecules. There are many different types of non-coding RNAs, including small nucleolar RNAs (snoRNAs), small interfering RNAs (siRNAs), microRNAs (miRNAs), and long non-coding RNAs (lncRNAs).
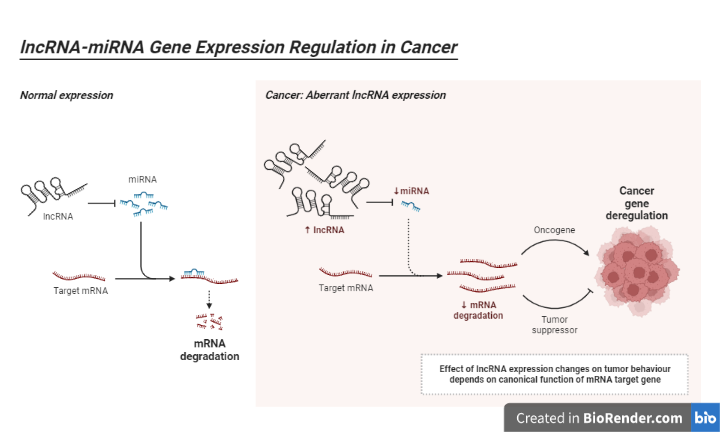
Fig: lncRNA-miRNA Gene Expression Regulation in Cancer
Basically, they are classified based on their length. Small noncoding RNAs are less than 200 nucleotides in length and contain both siRNAs and microRNAs (miRNAs). Long noncoding RNAs are transcripts with more than 200 nucleotides (lncRNAs). Almost every biological process involves microRNAs. lncRNAs also influence gene and protein levels.
Non-coding RNAs are often transcribed from DNA sequences that do not contain protein-coding genes, but they can also be transcribed from introns or from within protein-coding genes. Non-coding RNAs play important roles in many aspects of biology, and they are involved in a variety of human diseases.
Modes:
There are two main types of RNA splicing: constitutive splicing and alternative splicing.
Constitutive splicing:
Constitutive splicing is a type of RNA splicing that occurs in all cells and results in the production of a single protein from a gene. This type of splicing involves the removal of all introns from the primary RNA transcript, leaving only exons.
During constitutive splicing, the spliceosome recognizes specific sequences at the ends of introns and removes the intron from the primary transcript. The spliceosome then brings the ends of the exons together and joins them, creating a continuous coding sequence. This mature RNA molecule is then transported out of the nucleus and into the cytoplasm, where it can be translated into a protein.
Constitutive splicing is important because it allows cells to produce functional proteins that are essential for their normal function. It is also the most common type of RNA splicing, as most genes are transcribed and spliced in this way.
Alternative splicing:
Alternative splicing is a type of RNA splicing that allows cells to produce multiple different proteins from a single gene by including or excluding different exons. This process can produce different isoforms of a protein that can have different functions or be expressed in different tissues. Alternative splicing is more common in eukaryotes than in prokaryotes.
There are several mechanisms that can be involved in alternative splicing, including exon skipping, where an exon is skipped during splicing and not included in the mature RNA molecule, and alternative 5′ splice site selection, where the spliceosome uses a different 5′ splice site to remove an intron.
Alternative splicing is regulated by a variety of factors, including transcriptional and post-transcriptional regulators, as well as the specific cell type and tissue in which it occurs. This regulation allows cells to produce the correct proteins at the correct time and in the correct place.
It is an important process that allows cells to generate a diversity of proteins from a limited number of genes. It is also an important mechanism for generating protein diversity and regulating gene expression in eukaryotes.
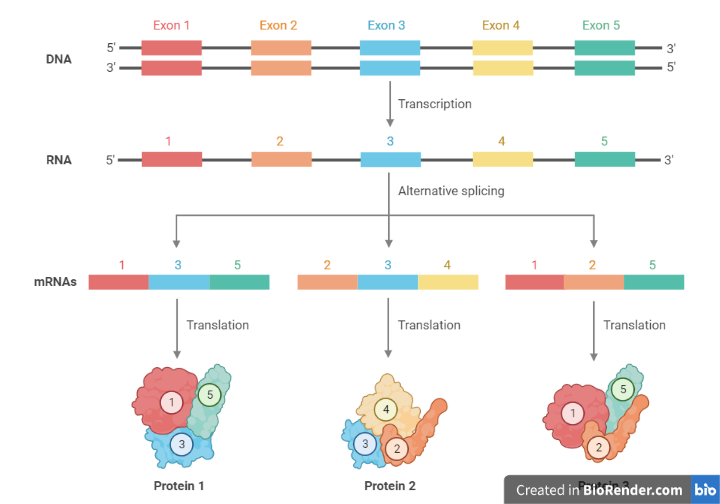
Fig: Alternative splicing
Different Types of Alternative Splicing:
Alternative splicing is a process by which different combinations of exons from a single gene can be spliced together to produce multiple different mRNA molecules, which can then be translated into different proteins with different functions. Alternative splicing allows for the production of multiple different proteins from a single gene and plays a key role in the regulation of gene expression. There are several different types of alternative splicing.
Cassette exon splicing: This occurs when an exon is either included or excluded in the final mRNA molecule, but all other exons remain unchanged.
Alternative 5′ splicing: This occurs when the start site of the mRNA molecule is changed, resulting in a different 5′ untranslated region (UTR).
Alternative 3′ splicing: This occurs when the stop site of the mRNA molecule is changed, resulting in a different 3′ UTR.
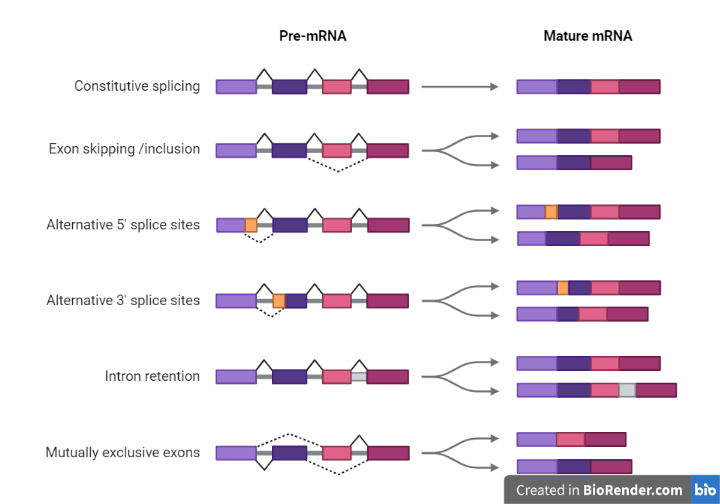
Fig: Different types of alternative splicing
Intron retention: This occurs when an intron is not removed from the transcript during splicing and is present in the final mRNA molecule.
Alternative promoter usage: This occurs when different promoters within a gene are used to initiate transcription, resulting in the production of mRNA molecules with different 5′ UTRs.
Process:
The process of RNA splicing occurs in several steps in which the primary RNA transcript is synthesized from DNA by the enzyme RNA polymerase. This transcript contains both exons (coding sequences) and introns (non-coding sequences). Furthermore, the primary transcript is transported into the nucleus, where splicing occurs.
The spliceosome, a complex of proteins and small RNA molecules, recognizes specific sequences at the ends of introns and removes the intron from the primary transcript.
The spliceosome brings the ends of the exons together and joins them, creating a continuous coding sequence. The mature RNA molecule is then transported out of the nucleus and into the cytoplasm, where it can be translated into a protein.
Recognition of introns: The spliceosome recognizes the introns in the pre-mRNA molecule based on specific sequences at the beginning and end of the introns called the 5′ splice site and the 3′ splice site, respectively.
Formation of the spliceosome: The spliceosome is assembled on the pre-mRNA molecule at the 5′ splice site. It consists of small nuclear ribonucleic acid (snRNA) molecules and proteins.
Cleavage of the phosphodiester bond: The spliceosome cuts the phosphodiester bond between the first nucleotide of the intron and the last nucleotide of the preceding exon. This releases the intron from the pre-mRNA molecule.
Formation of the lariat structure: The free 3′ end of the intron is then covalently bonded to a specific adenine nucleotide at the 5′ end of the intron, forming a lariat structure.
Cleavage of the phosphodiester bond: The spliceosome then cuts the phosphodiester bond between the last nucleotide of the intron and the first nucleotide of the following exon. This releases the intron from the pre-mRNA molecule.
Joining of exons: The spliceosome then joins the two exons together by forming a phosphodiester bond between the last nucleotide of the preceding exon and the first nucleotide of the following exon.
Release of the spliceosome: The spliceosome is then released from the mRNA molecule, leaving behind the mature mRNA molecule, which can be translated into a protein.
Spliceosome:
The spliceosome is a complex molecular machine composed of a myriad of proteins and small nuclear ribonucleic acid (snRNA) molecules that is responsible for the splicing of pre-mRNA (precursor mRNA). Splicing is the process by which introns, or non-coding regions, are removed from the pre-mRNA molecule and the remaining exons are joined together to form mature mRNA. The spliceosome recognizes specific sequences on the pre-mRNA called splice sites and then cuts out the introns and joins the exons together.
The spliceosome is composed of five small nuclear ribonucleoproteins (snRNPs) called U1, U2, U4, U5, and U6. Each snRNP contains one or more snRNA molecules and a set of proteins. The proteins in the spliceosome help to stabilize the snRNAs and facilitate the splicing reaction. The major spliceosome is composed of five snRNPs, designated U1, U2, U4/U6, and U5. Each snRNP contains a small nuclear RNA molecule and a set of proteins. The U1, U2, U4/U6, and U5 snRNPs are organized into a complex called the “core” spliceosome, which is responsible for recognizing and splicing introns. The core spliceosome is activated by the binding of additional proteins and snRNPs, which form the “activated” spliceosome. The spliceosome undergoes a series of conformational changes during the splicing reaction, which involve the rearrangement of the snRNPs and proteins that make up the complex.
Alternative Splicing and fusion protein:
A fusion protein is a protein that is created from the joining, or fusion, of two separate proteins. This can occur through alternative splicing, or it can be the result of a genetic mutation that leads to the fusion of two genes. Fusion proteins can have a variety of functions, depending on the proteins that have been fused together. They are often used in research and have potential therapeutic applications.
Fusion proteins can be created artificially in the laboratory by combining two proteins through genetic engineering techniques. They can also occur naturally as a result of genetic mutations that cause two genes to be fused together. Fusion proteins can be used for a variety of purposes, including as diagnostic markers, as targets for drug discovery, and as therapeutic agents. Some examples of fusion proteins include fusion oncoproteins, which are involved in cancer development, and receptor-ligand fusion proteins, which are used in drug delivery.
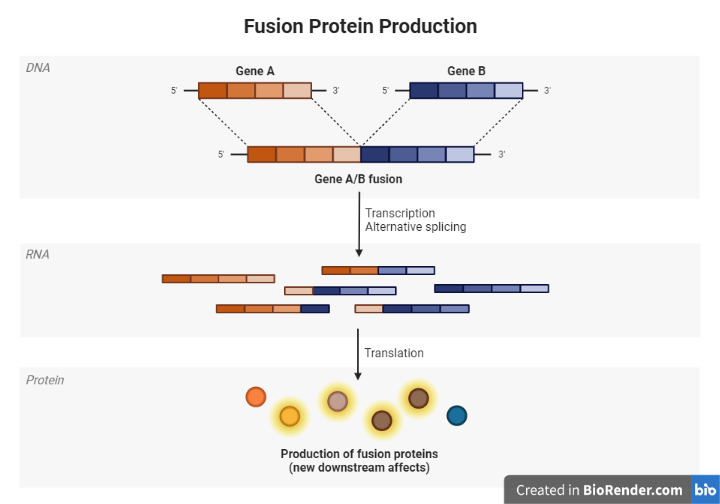
Fig: Alternative Splicing and fusion protein
Errors:
Errors in splicing can occur when there are mistakes in the splicing process, such as the incorrect removal of introns or the inclusion of non-coding DNA in the final mRNA product. The splicing process itself is a complex process that involves the recognition and removal of introns, or non-coding sections of DNA, from pre-mRNA molecules. This process is carried out by a complex of proteins and ribonucleic acids (RNAs) called the spliceosome.
During splicing, the spliceosome recognizes specific sequences at the ends of introns, called splice sites, and removes the introns from the pre-mRNA molecule. The remaining exons are then spliced together to form the final mRNA molecule. Errors in splicing can occur when the spliceosome makes mistakes during this process, such as by including non-coding DNA in the mRNA product or by failing to remove all of the introns from the pre-mRNA molecule.
Errors in splicing can be caused by a variety of factors, including genetic mutations, environmental factors, and changes in the expression of splicing regulatory proteins. They can also be influenced by the presence of certain diseases or conditions, such as cancer or neurodegenerative disorders. In some cases, errors in splicing can be corrected through the use of splicing modulators, which are drugs that target splicing regulatory proteins and can alter the splicing process to produce more normal mRNA and protein products.
Importance:
It is an essential process that occurs during gene expression in order to produce functional proteins.
This allows a single gene to give rise to multiple protein isoforms, which can have different functions and play a role in the diversity of proteins that are produced in an organism.
It is important for the proper expression of genes and the production of functional proteins.
Errors in splicing can lead to the production of abnormal or non-functional proteins, which can have a variety of consequences, depending on the protein that is affected.
Splicing is regulated by a complex network of proteins and RNA molecules, and changes in splicing patterns can be influenced by a variety of factors, including genetic mutations, environmental factors, and disease states.
Splicing modulators, which are drugs that target splicing regulatory proteins, have the potential to alter the splicing process and correct errors in splicing. These drugs are being developed for use in the treatment of various diseases.
References:
- van den Hoogenhof, M.M., Pinto, Y.M. and Creemers, E.E., 2016. RNA splicing: regulation and dysregulation in the heart. Circulation research, 118(3), pp.454-468.
- Ghosh, S. and Sinha, J.K., 2017. Size of Introns.
- https://www.nature.com/scitable/topicpage/rna-splicing-introns-exons-and-spliceosome-12375/
- Mattick, J.S. and Makunin, I.V., 2006. Non-coding RNA. Human molecular genetics, 15(suppl_1), pp.R17-R29.
- Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011 Jul 1;3(7):a003707.
