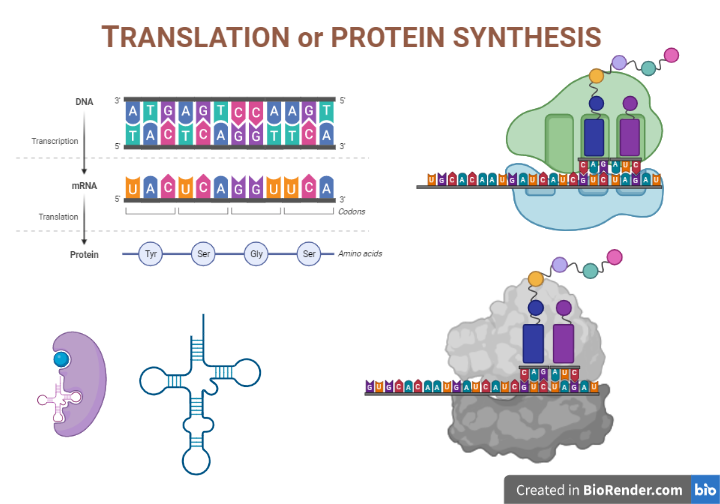
Introduction:
Translation is a molecular event of central dogma in which protein is synthesised from the sequence of the m-RNA. It is the process of decoding the genetic code contained within a messenger RNA (mRNA) molecule to produce a specific sequence of amino acids in a polypeptide chain. The process of is known as translation. In fact, protein synthesis encompasses the translation of mRNA’s nucleotide base sequence into amino acid sequence language.
Occurrence:
The translation mechanism is located in a specialized organelle called the ribosome in every cell. In eukaryotes, mature mRNA molecules must translocate from the nucleus to the cytoplasm to find ribosomes. In prokaryotic species, on the other hand, ribosomes can connect to mRNA while it is still being transcribed. Translation begins at the 5′ end of the mRNA while the 3′ end is still connected to DNA in this condition.
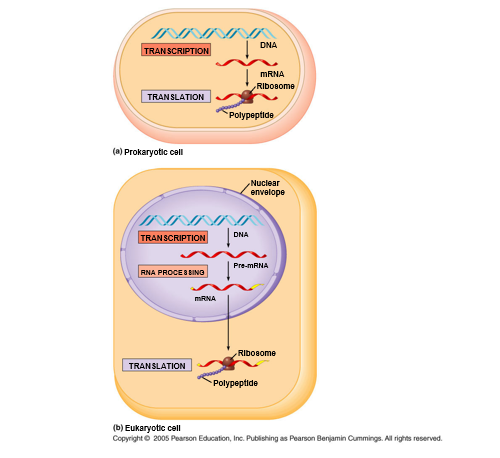
Fig: The Central Dogma in prokaryotic versus eukaryotic cells
Requirement of the components:
The key components required for translation are:
- Amino Acid
- Ribosomes
- Messenger RNA (mRNA)
- Transfer RNAs (tRNAs)
- Energy sources
- Protein factors
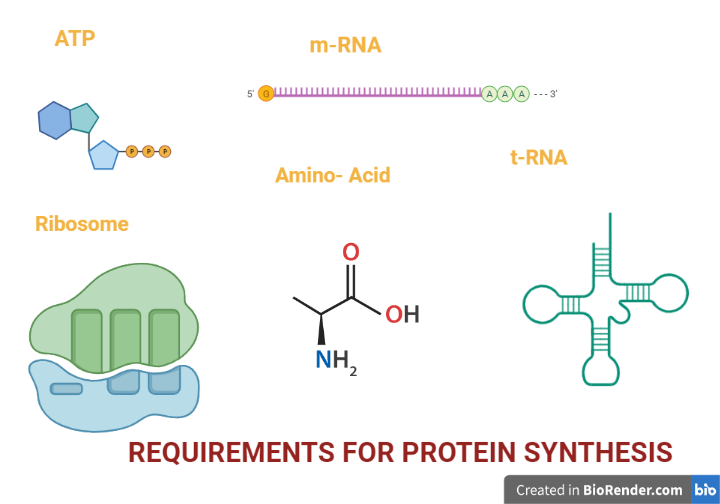
Fig: Requirements for protein synthesis
During translation, a codon is a three-base sequence found in mRNA that encodes a specific amino acid. Every tRNA molecule has an anticodon that is complementary to the mRNA codon, and the associated amino acid is at the other end (acceptor arm). As a result, tRNA molecules are in responsible for transporting amino acids to the ribosome in the proper order, ready for polypeptide synthesis.
A number of protein factors are required for initiation, elongation and termination of protein synthesis. The complexity of the protein factors is high in eukaryotes with respect to prokaryotes.
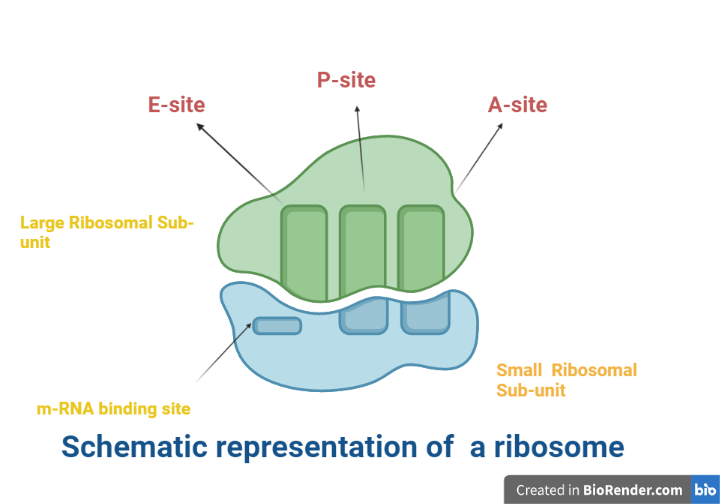
Fig: Schematic representation of a ribosome
Stages of protien sythesis:
Activation of Amino Acids
An enzymeaminoacyl-tRNA synthetase is responsible for activation of an amino acid for protein synthesis which involved two steps.
As stated previously, the energy of ATP hydrolysis is used to form a high-energy bond between each amino acid and its tRNA molecule. Two significant chemical conditions must be fulfilled in order to synthesize a polypeptide with a defined sequence:
- To facilitate the establishment of a peptide bond, each amino acids carboxyl group must be activated
- Each new incoming amino acid must be linked to other amino acid which will be encounter as per the codon in the mRNA.
In the initial phage, attachment of right amnio acid to a correct tRNA is crucial for protein synthesis which takes place in the cytosol. Each of the 20 amino acids is covalently attached to a specific tRNA to form aminoacyl tRNA. This reaction is possible using Mg2 dependent activating enzymes known as aminoacyl- tRNA synthetases at the expense of ATP energy. The attachment of amnio acid to a tRNA is said to be “charged.”
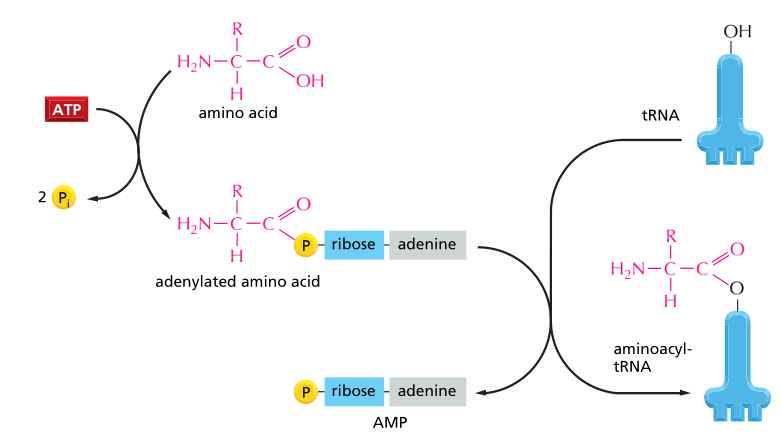
Fig: Activation of amino acid
Initiation:
Eukaryotes
In eukaryotes, the initiation of translation is a complicated process that involves at least ten eukaryotic initiation factors (eIFs).
The 80S ribosome breaks down into 40S and 60S subunits. Two initiating factors namely eIF-3 and eIF-1A (anti-association factor) bind to the newly formed 40S subunit, and thereby block its reassociation with 60S subunit. It led to formation of 43S preinitiation complex, 48S initiation complex and followed by 80S initiation complex in the involvement of GTP, met-tRNAi and several initiation factors.
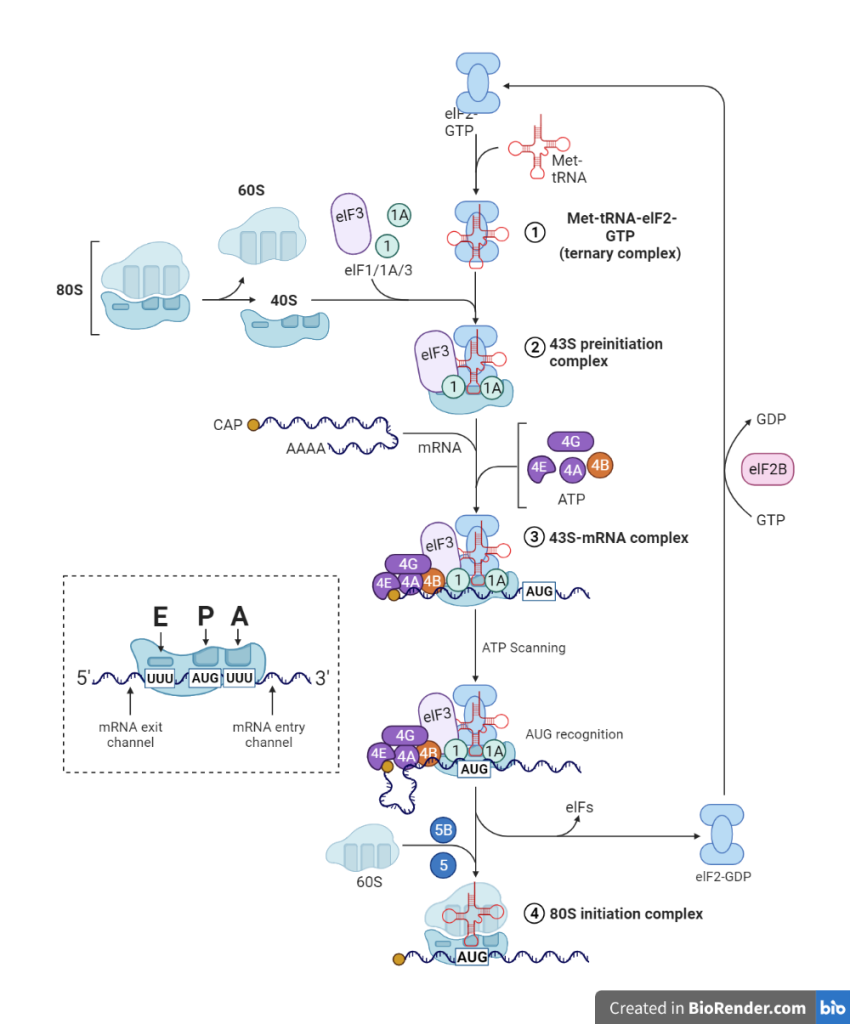
Fig: A diagrammatic representation of initiation of protein biosynthesis (translation) in eukaryotic cells
Prokaryotes
The formation of translation initiation complex in prokaryotes is less complicated compared to eukaryotes.
The 30S ribosomal subunit is associated to a ternary complex of IF-2, formyl met-tRNA, and GTP via initiation factor 3 (IF-3). Another initiation factor, IF-I, helps to form the preinitiation complex. The Shine-Dalgarno sequence is used to recognize the start codon AUG. In prokaryotes, a 50S ribosome unit is now bound with a 30S unit to form a 70S initiation complex.
Complementary base-pairing between the anticodon on the associated tRNA molecule and the next codon on the mRNA chain determines each amino acid introduced to the expanding end of a polypeptide chain. The codon selects the precise amino acid to be added to the expanding polypeptide chain since only one of the various varieties of tRNA molecules in a cell may base-pair with each codon.
Elongation:
Elongation requires elongation factors, which are cytosolic proteins. The hydrolysis of GTP as each residue is added to the developing polypeptide facilitates the binding of each incoming aminoacyl-tRNA and the movement of the ribosome along the mRNA.
The length of the nascent polypeptide is extended by the covalent attachment of consecutive amino acid units, which are transported to the ribosome and correctly positioned by their tRNA, which base-pairs to the respective codon in the mRNA.
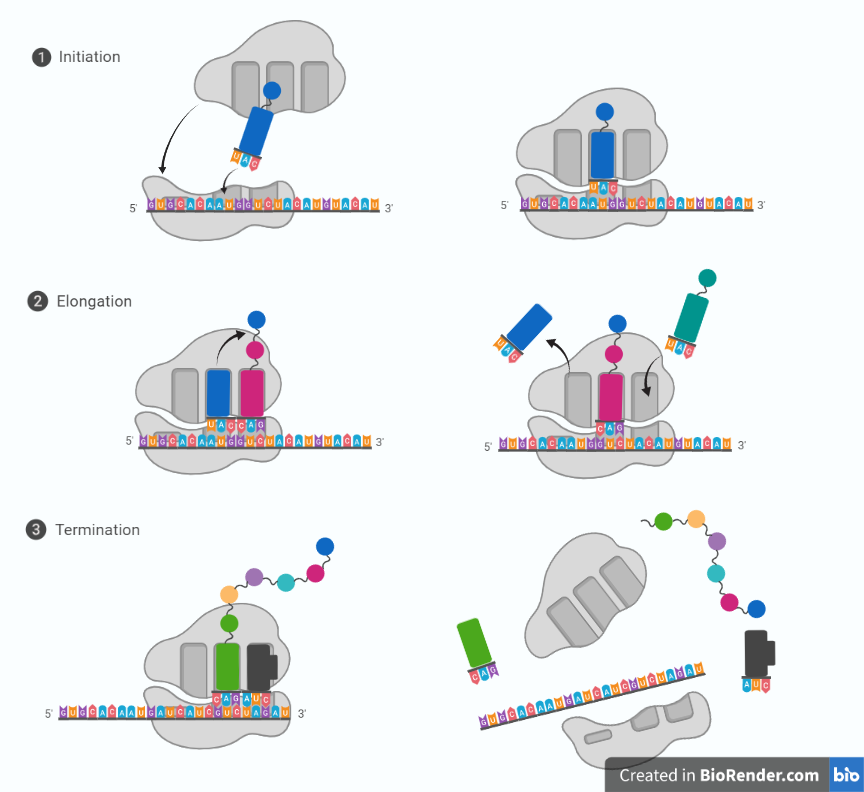
Fig: Steps in translation
In step 1, A non-charged tRNA (naked tRNA, that don’t attach amino acid) molecule dissociates from the E-site and as an aminoacyl-tRNA molecule binds to a vacant A-site on the ribosome.
In step 2, a new peptide bond is formed.
In step 3, the large subunit translocates relative to the small subunit, leaving the two tRNAs in hybrid sites: P on the large subunit and A on the small, for one; E on the large subunit and P on the small, for the other.
In step 4, the small subunit translocates carrying its mRNA a distance of three nucleotides through the ribosome.
Step 5: This “resets “the ribosome with a fully empty A-site, ready for the next aminoacyl-tRNA molecule to bind. As indicated, the mRNA is translated in the 5to-3 directions, and the N-terminal end of a protein is made first, with each cycle adding one amino acid to the C-terminus of the polypeptide chain.
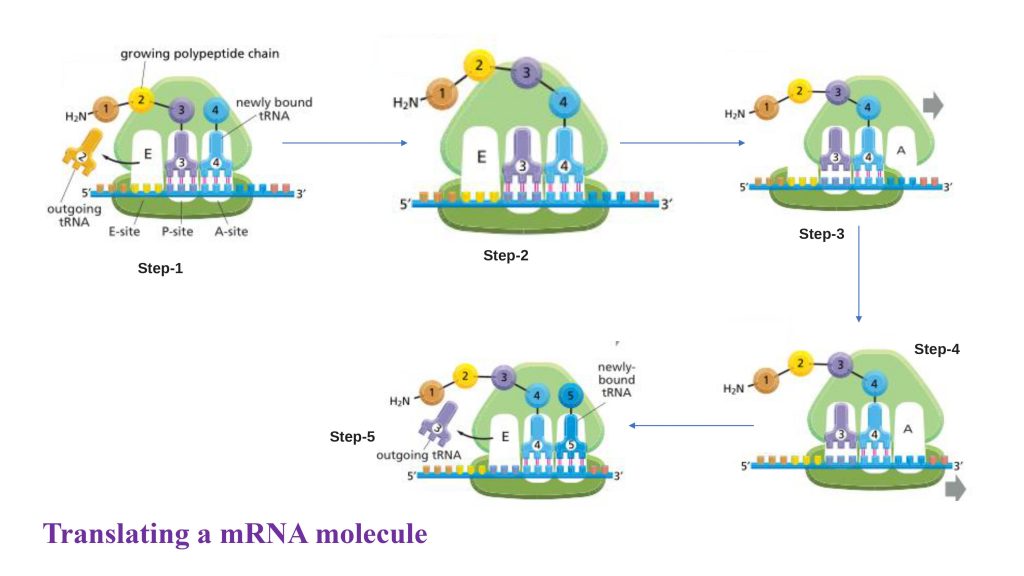
Fig: Translating an mRNA molecule
Termination and Release:
A termination codon in the mRNA indicates the end of the polypeptide chain. The release of the new polypeptide from the ribosome is facilitated by proteins known as release factors.
The three termination codons employed at the end of a protein-coding sequence in mRNA are UAA, UAG, and UGA. None of the tRNAs recognize these codons. As a result, one of a range of proteins known as release factors binds to these tRNAs and aids in the release of mRNA from the ribosome and subsequent ribosomal dissociation.
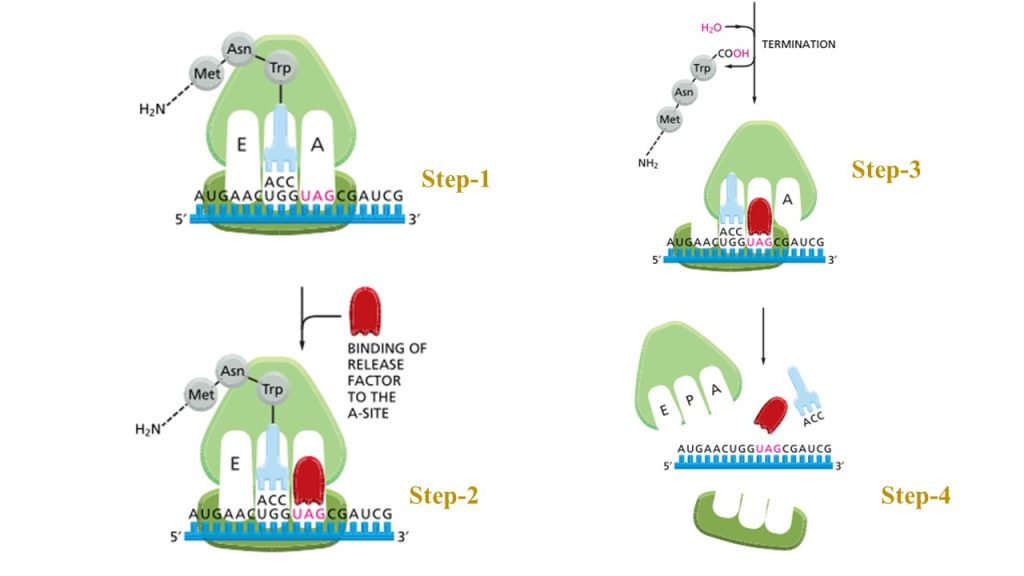
Fig: The final phase of protein synthesis
Folding and Posttranslational Processing:
Protein posttranslational modification (PTM) refers to the reversible or irreversible chemical modifications that proteins may undergo after translation. In other words, PTMs involve folding of new polypeptide into its proper three-dimensional conformation in order to achieve its biologically active form.
Enzymatic breakage of peptide bonds to covalent additions of specific chemical groups, lipids, carbohydrates, or even whole proteins to amino acid side chains are all examples of chemical changes. After biosynthesis, these chemical alterations of a polypeptide chain broaden the spectrum of amino acid structures and characteristics, resulting in a greater diversity of protein structures and functions.
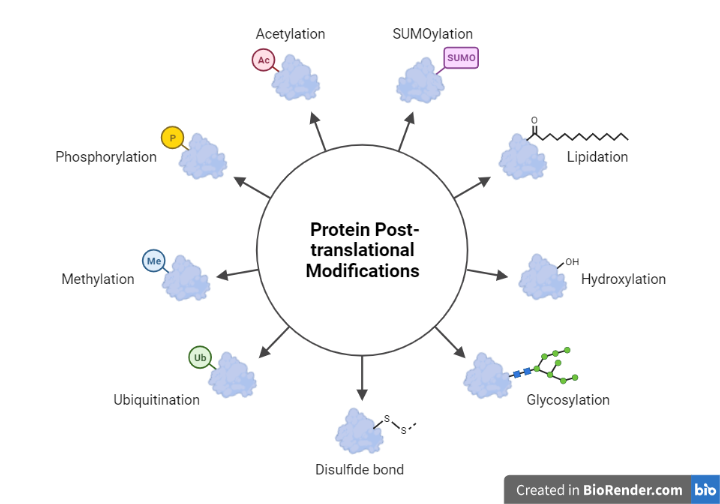
Fig: Post Translation modification
Comparing Eukaryotic and Prokaryotic Translation:
In both prokaryotes and eukaryotes, the translation process is relatively similar. The genetic code is largely same, despite the utilization of distinct elongation, initiation, and termination factors. As previously stated, transcription and translation occur simultaneously in bacteria, and mRNAs have a short lifespan. In eukaryotes, however, mRNAs have extremely variable half-lives, are sensitive to changes, and must transit the nucleus to be translated; these many processes provide additional possibilities to fine-tune gene expression levels.
Protein Synthesis Inhibitors:
A protein synthesis inhibitor is a substance that prevents or inhibits cell growth or proliferation by interfering with the mechanisms that lead to the production of new proteins. They are medications that prevent bacteria from producing proteins and thereby preventing them from multiplying. Protein synthesis inhibitors, such as erythromycin, tetracycline, chloramphenicol, and aminoglycosides, are another important class of clinically beneficial antibacterials. They only interact with the 70S bacterial ribosome particle, leaving the 80S eukaryotic ribosome particle alone.
Diphtheria toxin (DT) is a bacterial toxin having intracellular activity that has been extensively characterized. Diphtheria symptoms are caused by toxigenic strains of Corynebacterium diphtheriae.
The mechanism of action of diphtheria toxin has been thoroughly studied. It kills cells by blocking eukaryotic protein synthesis. Elongation factor (EF-2) is a protein synthesis factor that is inactivated by this toxin.
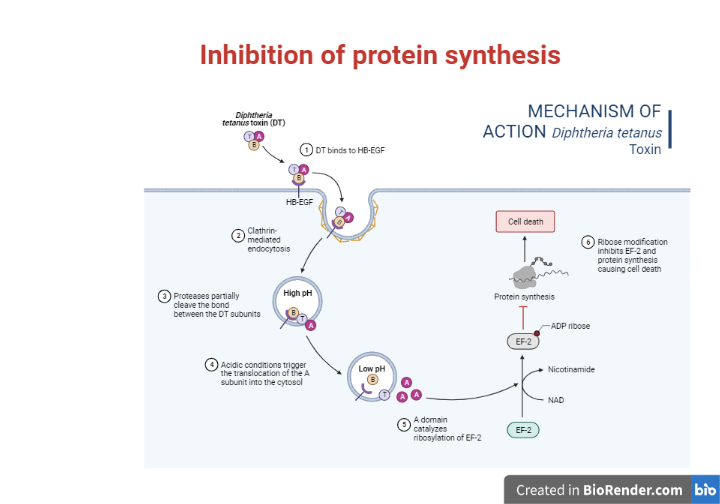
Fig: Inhibition of protein synthesis
References:
- Clancy, S. and Brown, W., 2008. Translation: DNA to mRNA to. Nature Education.
- Alberts, B., Johnson, A., Walter, P., Lewis, J., Raff, M. and Roberts, K., 2008. Molecular cell biology. New York: Garland Science.
- Satyanarayana, U., 2021. Biochemistry, 6e-E-book. Elsevier Health Sciences.
- David, L., Nelson, D.L., Cox, M.M., Stiedemann, L., McGlynn Jr, M.E. and Fay, M.R., 2000. Lehninger principles of biochemistry.