Introduction
Chromosomal analysis is a widely used technique in diagnosing chromosome instability and rearrangements that contribute to genetic diseases and cancer.
Chromosomes can be isolated from cells of living tissues such as blood lymphocytes, skin fibroblasts, amniocytes, placenta, bone marrow, and tumor specimens. Chromosomes are studied at the metaphase stage of mitosis, when they are most compacted and hence more visible. Phytohaemagglutinin (PHA), a lectin derived from the red kidney bean, is a powerful mitogen for human T-cells (thymus-derived small lymphocytes) When PHA is added in vitro to whole blood, mitotic cells can be found after 48 h, with a peak mitotic index at ~64-72 hr. 72 hours after the addition of PHA to the culture, about 45% of cells are in S phase.
This reflects the maximum of mitotic activity and is the ideal time to collect for chromosomal for investigations. When studying B cells, other mitogens such as pokeweed (1-10 g/ml) may be added. The first step of the chromosome isolation technique involves the disruption of the spindle fibers by incubation with Colcemid, to prevent the cells from proceeding to the subsequent anaphase stage. The cells are then treated with a hypotonic solution and preserved in their swollen state with Carnoy’s fixative. The cells are then dropped on to slides and can then be utilized for a variety of procedures. G-banding involves trypsin treatment followed by staining with Giemsa to create characteristic light and dark bands. The same procedure to isolate chromosomes can be used for the preparation of cells for procedures such as fluorescence in situ hybridization (FISH), comparative genomic hybridization (CGH), and spectral karyotyping (SKY).
Note:
Other mitogens used to excite B cells include cytochalasin B, Epstein-Barr virus (EBV), protein A, and lipopolysaccharides (LPS). Phytohemagglutinin (PHA) is a lectin (a type of protein) that is found in certain plants, such as kidney beans, red kidney beans and broad beans. PHA has the ability to agglutinate (clump together) red blood cells and has been used in medical research as a mitogen, a substance that stimulates cell division.
Principle:
Blood lymphocytes are stimulated by the mitogen phytohemagglutinin (PHA) to rapidly grow and divide such that three days of culturing will yield late prophase or early metaphase cells of sufficient quantity and quality for clinical analysis. Colcemid inhibits the formation of the spindle fibers. When added to the stimulated culture, it causes mitotically active cells to be arrested in early metaphase. The use of methotrexate and thymidine to synchronize the cultures allows a significant number of cells to enter early metaphase near the time of harvest. By intercalating with the DNA and preventing chromosome condensation, ethidium bromide can also yield chromosomes with greater length and resolution.
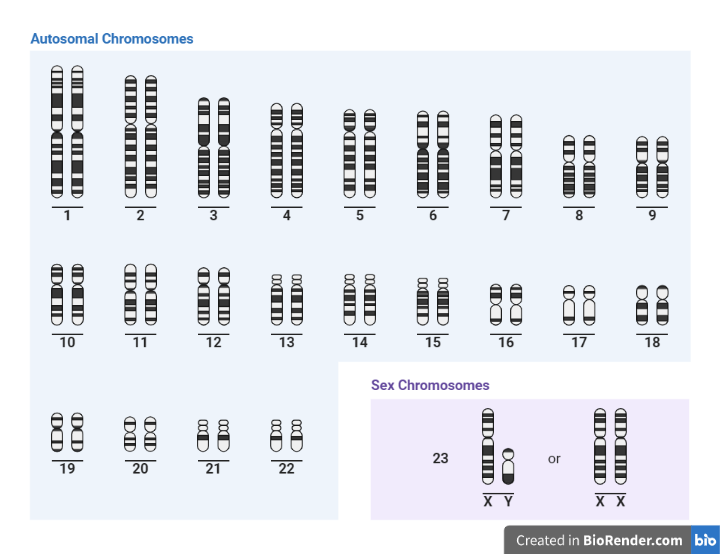
Fig: Karyotyping
Sample:
Blood (5 mL)
Collect at least 1 ml of whole blood in a green top sodium heparin tube. Use within 3 days after collection. Store blood at room temperature until ready to use.. Lithium heparin or EDTA are toxic to cells and should never be used.
Reagents:
- Carnoy’s fixative (freshly prepared): 3:1 ratio of methanol: glacial acetic acid
- Colcemid (10 μg/ml)
- Ethidium bromide (1 μg/mL)
- Fetal bovine serum
- Gurr’s buffer solution: Dissolve one Gurr pH 6.8 buffer tablet in 1 L of H2O. Store at 4°C.
- Hypotonic solution (37 °C preheated, 0.075 M KCl)
- Mounting medium
- Phytohaemagglutinin
- Trypsin solution (5%)
- Wright stain, Giemsa stain or Leishman stain
Equipments:
- Centrifuge (benchtop, 200g)
- Coverslips
- Gloves
- Hair dryer
- laminar flow
- Incubator preset to 37°C, 5% CO2, 100% humidity
- Karyotyping station
- Microscope
- Immersion oil
- Photomicroscope equipped with 10X and 100X objectives
- Pipettes (disposable)
- Slide warmer, hotplate, or oven preset to 56°C-60°C
- Microscopic slides
- Staining jars (Coplin)
- Tubes (polypropylene, sterile, 10-mL)
- Vortex mixer
- Water bath preset to 37°C
Method:
Lympocyte Culture
Working under the tissue culture hood and using sterile technique, aliquot complete RPMI into three polypropylene tubes, 4.5 ml per tube
Harvesting can be done at various times with numerous cultures, and cultures with the greatest number of high-quality metaphases can be used for analysis.
- Add 0.5, 0.45, or 0.4 ml of whole blood to each tube
- Add 0.1 ml of PHA to each tube
- Tighten the caps. Mix by inverting several times. Loosen the caps sufficiently and tilt the tube at 45° in order to promote air exchange to allow for exchange of gases
Tilting culture tubes to 45° increases gas exchange and prevents cells from packing too densely at the bottom.
- Place in the incubator for 64-72 h at 37°C with 5% CO2.
- Each tube should have 50 μL of ethidium bromide added two hours prior to harvest.
Ethidium bromide inhibits chromosome contraction, facilitating the analysis of longer mes with higher band resolution. Depending on the objective of the analysis, ethidium bromide may not be needed.
- Forty-five minutes before harvesting, add 50 μl of colcemid solution to each tube. Return the tubes to the incubator.
The addition of the mitotic inhibitor (Colcemid) accumulates metaphases by preventing the formation of the spindle apparatus. Mitotic inhibitors also cause chromosome contraction.
Cell Harvesting
- Remove the tubes from the incubator. Centrifuge at 200g for 8 min.
- Carefully remove as much of the supernatant as possible with a Pasteur pipette, taking care not to disturb the white cell layer above the red cell pellet.
- Mix the red and white cell layers with a Pasteur pipette or vortex mixer.
- Slowly add 1 ml of hypotonic solution (prewarmed to 37°C) dropwise, mixing well with the cell suspension. Continue to add hypotonic solution to a final total volume of 10 ml.
As the volume in the tube increases, the rate at which the hypotonic solution is added can be increased gradually. Chromosome spreading will grow with more time, but since this treatment is a hypotonic “shock,” adding more hypotonic solution will have a greater impact.
- Incubate in a water bath at 37°C for 10 min.
- Add 1 mL of freshly prepared Carnoy’s fixative to each tube. Cap the tubes. Mix by inverting.
The purpose of this procedure is to gradually lower the pH of the cells in order to prepare them for the subsequent fixation processes. Additionally, it starts the process of removing the cellular debris produced by the lysis of any remaining red blood cells.
- Centrifuge at 200g for 8 min. Using a Pasteur pipette, carefully remove the supernatant while being careful not to disrupt the cell pellet.
- Break up the pellet by agitation. Dropwise add 0.5 mL Carnoy’s fixative, mixing after each drop. Continue to add fixative to a final total volume of 5 ml.
As the volume in the tube increases, the rate at which the fixative is added can be increased gradually. This technique gradually lowers the pH of the cells, preparing them for the subsequent fixation stages. It also lyses leftover red blood cells and starts the process of removing the cellular debris that results.
Repeat Steps 1 and 2 twice more. Resuspend the final pellet by dropwise addition of Carnoy’s fixative to a final total volume of ~1 ml.
In unfavorable harvests, longer fixation will frequently boost chromosomal spreading. Keeping the suspension overnight at 4◦C can improve the quality of the preparation or can be done for scheduling reasons. Suspensions should be kept in polypropylene tubes containing plenty of fixative. Polystyrene tubes will react with fixative and should not be used.
Slide Preparation
Flooding the slide with fixative immediately following dropping the cells on to the slide will also help chromosomes to spread. Temperature and humidity, which affect how fast the cell suspension dries on to the slide, are other factors which affect chromosome spreading. These can be controlled by preparing slides in an environment with approximately 50% humidity and a temperature of about 20-25 °C, and/or by placing slides on a slide warmer for faster drying.
- Incubate the slides at least 10 minutes (or up to 3 days) on a 56°C-60°C slide warmer, hot plate or oven
- Examine the slide under a phase contrast microscope to evaluate the quality of the preparation and the extent of chromosome spreading. If the quality is satisfactory, prepare additional slides using the same technique.
G-banding (GTW Banding)
- Prepare the staining jars:
- Rinse the slide to be stained in the first jar (first rinse) for 1-2 min.
- Transfer the slide to the second jar (working trypsin solution) for ~30 sec.
The time required for trypsin digestion varies. Trypsin is a proteolytic enzyme that denatures euchromatic histones in DNA regions, resulting in increased transcriptional activity. These areas will emerge as light bands after Giemsa staining. Heterochromatin (highly condensed chromatin with little or no transcriptional activity) will have a major amount of its histones shielded from trypsin and will thus stain darkly after Giemsa staining. For best results, trypsin should always be kept cold and slide should remain hot. Once hot slides are introduced into cold trypsin, thermal shock can deliver better results.
- Place the slide in the third jar (arresting solution) for ~1 min.
- Rinse the slide in the fourth jar (second rinse) for ~1-2 min.
- While the slide is the fourth jar, prepare a working stain solution by mixing 0.5 mL Wright stain with 2 mL of Gurr’s buffer per slide.
- Transfer the slide from the fourth jar to the horizontal slide support. Gently flood the slide with the stain. Stain for ~40-50 sec.
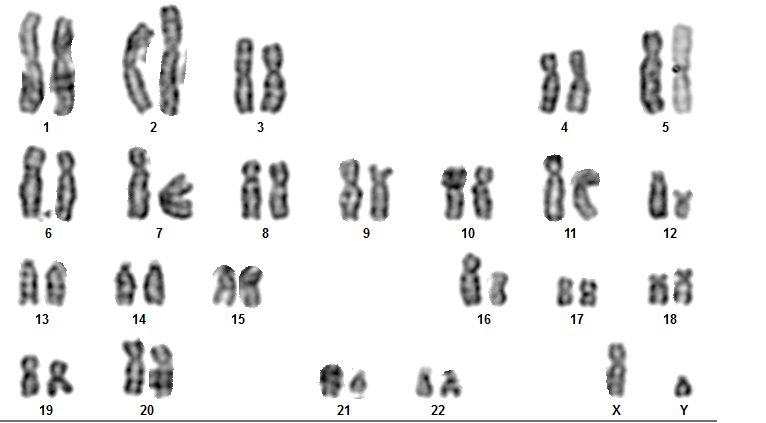
Fig: Karyotiping
The time required for staining can differ.
- Rinse off the stain with H2O. Dry the slide completely using a hair dryer.
- Place two drops of mounting medium on the slide. Make sure there are no air bubbles before carefully adding a coverslip.
- Allow the coverslip to dry for 2-3 min before examining the sample under the microscope.
Chromosome Analysis
- Identify individual well-spread and banded cells at low magnification.
- Analyze suitable preparations at high magnification
- For routine diagnostic cytogenetic analysis, examine at least 20 cells to help rule out mosaicism.
A much greater number of cells must be scored for chromosome breakage studies.
- Using an automated karyotyping station, digitally record high-resolution images of individual metaphase cells. Separate the chromosomes and arrange them into a karyotype. Alternatively, cells can be photographed, printed, and each chromosome cut from the picture and arranged into a karyotype.
Applications:
Diagnosis of genetic disorders: By analyzing the chromosomes of a person with a suspected genetic disorder, karyotyping can identify specific chromosomal abnormalities that are known to cause certain diseases.
Cancer diagnosis and treatment: Karyotyping can be used to analyze the chromosomes of cancer cells to identify any chromosomal abnormalities that may be associated with the cancer, which can help in determining the best course of treatment.
Stem cell research: Karyotyping can be used to evaluate the chromosomes of stem cells to ensure that they are normal and suitable for use in research and therapy.
Prenatal diagnosis: Karyotyping can be used to evaluate the chromosomes of a developing fetus in order to identify any chromosomal abnormalities that may indicate a genetic disorder or birth defect.
Assisted reproductive technology: Karyotyping can be used to evaluate the chromosomes of embryos produced through in-vitro fertilization (IVF) to identify which ones are most likely to lead to a successful pregnancy.
Limitations:
- Karyotyping can only detect changes in chromosome structure that are visible under a microscope.
- Karyotyping can only be performed on cells that are actively dividing, making it difficult to analyze tissues such as nerve cells.
- The process of growing and preparing cells for karyotyping can take several days.
- Karyotyping can only detect changes in the structure of chromosomes, it can’t detect changes in the DNA sequence.
- Low grade clonal rearrangements and/or the presence of submicroscopic or cryptic abnormalities may not be evident on conventional karyotyping.
References:
- Behrend, C., Hagh, J.K., Mehdipour, P. and Schwanitz, G., 2017. Human Chromosome Atlas: Introduction to diagnostics of structural aberrations. Springer.
- Calonje, E., Brenn, T., Lazar, A. and McKee, P.H., 2012. McKee’s pathology of the skin: with clinical correlations. Elsevier/Saunders.
- Tan, Q.K.G., 2021. Diagnosis of Genetic and Metabolic Conditions.
- Smith, K., 2006. Basic Cytogenetic Techniques: Culturing, Slide Making, and G Banding. In Cell Biology (pp. 381-385). Academic Press.
