Introduction to Protein Secretion in Bacteria:
Like all other organisms, bacteria have all the necessary cell structures that ensure their survival and proper functioning. The protective cell envelope can be two or three membranes, depending on the nature of the bacteria. Bacteria are protected from the environment by one or two membranes that prevent the movement of small molecules (e.g., ions and metals) and macromolecules (e.g., proteins and DNA) within the bacterial cell and between the environment and the bacteria. Bacteria synthesize proteins within their cells in the ribosome either for structural purposes (eg, membrane proteins of bacteria), synthesis of organelles, obtaining nutrients, and for coding virulence factors or export of proteins within other cell compartments or to the outside environment.
Of the variety of mechanisms employed by bacteria to transport proteins, the general secretory (Sec) pathway and the twin-arginine translocation (Tat) system are indispensable pathways that aid in protein translocation.
For proteins to be transported in an unfolded condition and undergo modification at their destination, the Sec pathway is essential. Tat system, on the other hand, transports folded proteins that are already modified and adjuncted with cofactors, if required, before secretion.
Sec Pathway- Mechanism and Function:
The Sec pathway mainly transports proteins across the membrane in an unfolded form. This pathway comprises a targeting moiety that targets proteins, a motor protein, and a channel that spans the membrane. This pathway is essential for translocating proteins that impart virulence to the bacteria. Both Gram-negative bacteria, such as Vibrio cholerae and Klebsiella pneumoniae, and Gram-positive pathogens like Staphylococcus aureus and Listeria exploit this system for protein secretion.
Proteins may be targeted to the translocase in two ways. The Sec pathway machinery recognizes a hydrophobic N-terminal leader sequence (SecB-specific signal sequence) on proteins that need to be transported and translocated actively using ATP and a proton gradient for energy, while proteins that remain in the cytoplasmic membrane contain a Signal Recognition Particle (SRP)-specific signal sequence.
Sec B Pathway- Mechanism and Function:
In numerous Gram-negative bacteria, proteins destined for secretion into the periplasm or outside the cell via the Sec pathway possess signal peptides specifically recognized by SecB. SecB interacts with the unfolded proteins and transfers them to SecA, an ATP-dependent motor protein that drives their passage through the membrane. The protease removes the signal sequence. After reaching the periplasm, the proteins fold into their functional forms and may subsequently be exported across the outer membrane through either the Type II or Type V secretion systems.
Signal Recognition Particle (SRP) pathway:
A second mechanism of protein secretion involves the translation of the synthesized protein along with its secretion through the SecYEG channel. The SRP pathway relies on the signal recognition particle (SRP). Once the SRP complex binds to its receptor, translation resumes and presumably provides the force to translocate polypeptides across the membrane.
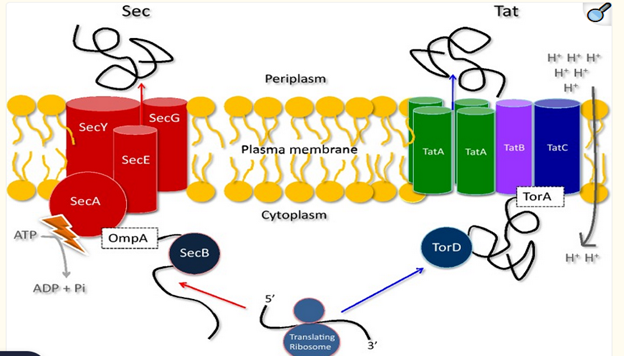
Source: Frain, K. M., Robinson, C., & van Dijl, J. M. (2019).
Fig: The core components and mechanisms of bacterial protein secretion via the Sec and Tat pathways, emphasizing their distinct roles in handling unfolded vs. folded protein cargo
Tat Pathway- Mechanism and Function:
The twin-arginine translocation (Tat) pathway is basically different from other bacterial secretion systems as it translocates folded proteins that are modified and incorporated with essential cofactors, if necessary, across biological membranes. The machinery responsible for Tat-mediated secretion is composed of three primary protein components: TatA, TatB, and TatC. These components operate in a coordinated manner to translocate target proteins across the inner membrane.
In Gram-positive bacteria, proteins exported via the Tat pathway are typically transported directly into the extracellular environment due to the absence of an outer membrane. TatA, TatB, and TatC are the three components that make up the Tat pathway in Gram-positive bacteria. Together, TatA and TatB form a single, multipurpose protein.
In Gram-negative organisms, the target proteins are first taken into the periplasmic space. They can then either stay in the periplasm or move farther over the outer membrane into the extracellular environment via additional export systems, most notably the Type II Secretion System (T2SS). Gram-negative bacteria, such as Escherichia coli, proteins containing specific N-terminal signal peptides with a conserved twin-arginine (RR) motif are first recognized by the TatBC complex, which serves as the initial docking site for the folded substrate. Upon successful recognition, TatA is recruited to the membrane-associated TatBC complex, where it assembles into a dynamic, pore-forming structure. TatA then mediates the actual translocation of the protein through the membrane by forming a transient aqueous channel that accommodates the passage of folded proteins.
References:
- Frain, K. M., Robinson, C., & van Dijl, J. M. (2019). Transport of Folded Proteins by the Tat System. The protein journal, 38(4), 377–388. https://doi.org/10.1007/s10930-019-09859-y
- Green, E. R., & Mecsas, J. (2016). Bacterial Secretion Systems: An Overview. Microbiology spectrum, 4(1), 10.1128/microbiolspec.VMBF-0012-2015. https://doi.org/10.1128/microbiolspec.VMBF-0012-2015
