Introduction:
Marker systems are techniques used to analyze gene transfer in an experimental organism.
In gene transfer research, a foreign gene, known as a transgene, is introduced into an organism through a process known as transformation. A typical issue for researchers is determining quickly and simply whether the transgenic has been taken up by the organism’s target cells.
A marker allows the investigator to determine whether the transgene was transferred, where it is placed, and when it is expressed.
A marker gene is a gene that is used to identify a certain cell or organism or to track the expression of a gene of interest. In molecular biology, a marker gene can be employed as a selectable marker to identify that a foreign gene has been effectively integrated into the genome of a host organism.
A marker gene in genetics can be used to identify a certain trait or phenotype. The presence or absence of particular marker genes on the surface of red blood cells, for example, determines the ABO blood type system.
Types:
Selectable marker genes
The gene of interest is introduced into a suitable vector and converted into host organisms in genetic engineering. Unfortunately, the transformation is only successful in competent host cells, and host cells may reject the uptake of foreign DNA.
To proceed with subsequent experiments, differentiation of transformed and non-transformed cells is required. As a result, researchers added a selectable marker gene to the vector to make it easier to distinguish transformed cells from non-transformed ones. A selectable marker is a DNA sequence, particularly a gene that can be used to identify altered cells. This indicator gene demonstrates a characteristic that lends itself to the artificial selection of transformed cells from non-transformed cells on a medium.
They are present in the vector alongside the target gene. The major portion of the time, the selection is based on the survival of the modified cells when cultured in a medium containing a harmful material (antibiotic, herbicide, antimetabolite).
A large number of selectable marker genes are available and they are grouped into three categories—
antibiotic resistance genes, antimetabolite marker genes and herbicide resistance genes
Antibiotic Resistance Genes
Antibiotic resistance genes serve as selective markers in several plant transformation systems. Despite the fact that plants are eukaryotic in origin, antibiotics can effectively limit protein biosynthesis in cellular organelles, particularly chloroplasts.
In general, the most often utilized antibiotic resistance genes include those that produce resistance to kanamycin, ampicillin, chloramphenicol, tetracycline, and gentamicin. The kanamycin resistance gene, for example, produces a protein that inactivates the antibiotic kanamycin, whereas the ampicillin resistance gene encodes a protein that degrades the antibiotic ampicillin.
Eg: Neomycin phosphotransferase II (npt II gene):
The npt II gene, which encodes the enzyme neomycin phospho-transferase II, is the most extensively used selectable marker (NPT II). This marker gene provides resistance to the antibiotic kanamycin. Transformants and plants generated from them can be tested with kanamycin solution, and resistant progeny can be selected.

Fig: Selectable Antibiotic resistance marker Genes
Antimetabolite Marker Genes
Antimetabolite marker genes encode enzymes that are sensitive to specific medications or analogs of natural metabolites, as opposed to antibiotic resistance genes, which provide resistance to specific antibiotics.
Eg: Dihydrofolate reductase (dhfr gene)
Methotrexate, an antimetabolite, inhibits the enzyme dihydrofolate reductase, which is encoded by the dhfr gene. A mutant dhfr gene in mice has been found that codes for this enzyme with a low affinity for methotrexate. When the dhfr gene is coupled with the CaMV promoter, it produces a methotrexate resistant marker that can be used to select transformed plants.
Herbicide Resistance Markers
These genes encode enzymes that confer resistance to certain herbicides, allowing transformed plants to live while non-transformed plants will die.
Eg: Enolpyruvylshikimate phosphate synthase (epsps/aroA genes):
Glyphosate, a herbicide, slows photosynthesis. It inhibits the action of enolpyruvylshikimate phosphate synthase, a crucial enzyme in the production of phenylalanine, tyrosine, and tryptophan.
Glyphosate-resistant mutant strains of Agrobacterium and Petunia hybrida have been found. Resistance to transgenic plants can be conferred by the genes epsps/aroA.
Reporter genes
Reporter genes are genes that are utilized in molecular biology studies to explore the regulation of gene expression. They are frequently introduced artificially into cells or organisms and can be used to monitor the activity of a gene or collection of genes. Reporter genes are usually fused with a gene of interest to form a chimera gene that can subsequently be expressed in cells or organisms. The reporter gene’s expression can subsequently be tracked and quantified using techniques like fluorescence microscopy, luminescence, or colorimetry. Reporter genes are an important tool for molecular biology research because they allow researchers to view and measure gene activity in real time, offering vital insights into gene control mechanisms.
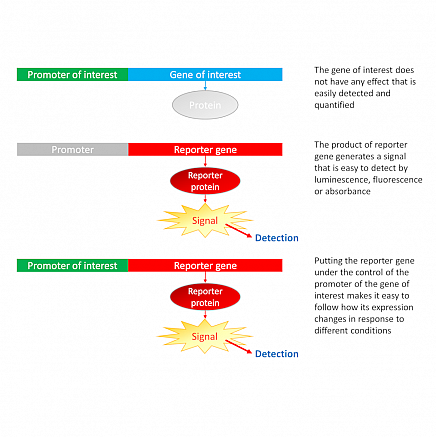
Source: https://www.berthold.com/en/
Fig: Reporter Gene
| Reporter Gene | Source | Mechanism | Detection Assay |
| Fluorescent Protein (GFP) | Aequorea victoria (jellyfish) | Emits green fluorescence when exposed to blue light | Fluorescence microscopy or flow cytometry |
| β-glucuronidase (GUS) | Escherichia coli (bacteria) | Catalyses the hydrolysis of glucuronides | Colorimetric assay using X-Gluc |
| Luciferase | Fireflies, bacteria, etc. | Catalyses the oxidation of luciferin to produce light | Luminescence assay using a luminometer |
| Discosoma sp. Red Fluorescent Protein (DsRed) | Discosoma sp. (coral) | Emits red fluorescence when exposed to blue or green light | Fluorescence microscopy or flow cytometry |
Fig: Selected reporter gene used in gene transfer in plants the source, mechanism and detection assays
Applications:
Monitoring gene expression
Marker genes are used to track the expression of a certain gene in real time. The activity of the gene can be observed and quantified by fusing the marker gene with the gene of interest using several detection methods such as fluorescence microscopy or colorimetry.
Identifying transformed cells
Marker genes can be used to identify cells that have effectively been transformed with an interest gene. Transformed cells can be selected and isolated from untransformed cells by inserting a selectable marker gene, such as antibiotic resistance, with the gene of interest.
Studying protein localization
Marker genes can be fused with a protein of interest to explore its subcellular localization. Researchers can determine the location of the protein of interest and investigate its function within the cell by viewing the position of marker gene.
Studying promoter activity
Marker genes can be used to investigate the activity of a promoter, which is a DNA region that regulates gene expression. The activity of the promoter can be observed and quantified by fusing the marker gene with the promoter.
References:
- Boulin, T., Etchberger, J.F. and Hobert, O., 2006. Reporter gene fusions. WormBook: The Online Review of C. elegans Biology.
- Miki, B. and McHugh, S., 2004. Selectable marker genes in transgenic plants: applications, alternatives and biosafety. Journal of biotechnology, 107(3), pp.193-232.Marker genes in gene therapy: A review by Karimian et al. (2019) in the journal Current Gene Therapy.
- Reporter genes in biotechnology: A review by Glaser et al. (2020) in the journal Biotechnology Advances.
- Marker genes in gene therapy: A review by Karimian et al. (2019) in the journal Current Gene Therapy.
