Introduction:
Transcription is the preliminary stage in DNA-based gene expression, in which the enzyme RNA polymerase copies a specific segment of DNA into RNA (primarily mRNA). It is the process of linking nucleotides (monomeric units for nucleic acids) to form a RNA molecules from DNA strands as templates in order to transmit genetic information. Transcription is the initial stage in gene expression, when information from a gene is used to build a functional product like a protein. Although the entire genome of DNA must be duplicated, only a small fraction of it is transcribed in response to developmental, physiological, and environmental changes.
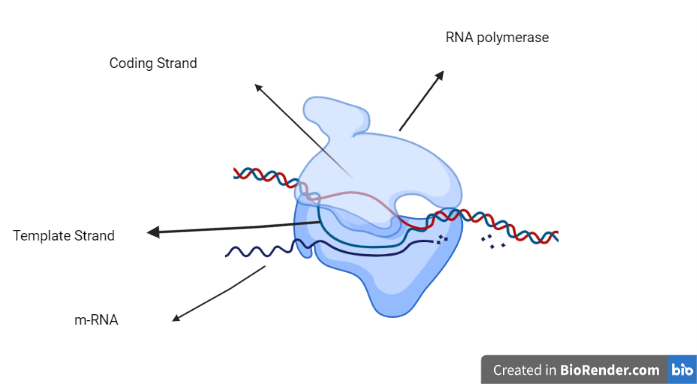
Structural genes are DNA segments that can be translated into RNA. The strand from which the RNA is actually transcribed is called the template strand. It’s also known as an antisense strand. The coding strand is the strand that specifies the amino acid sequence of the encoded protein by its base sequence. As a result, it is also known as the sense strand. An RNA polymerase reads a DNA sequence during transcription and creates a corresponding, antiparallel RNA strand known as a primary transcript.
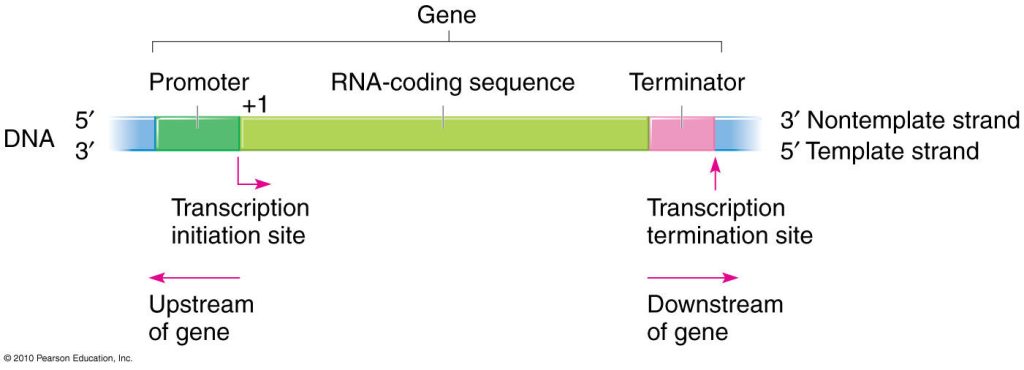
Fig: Model of Transcription unit
A transcription unit is a segment of DNA that connects the locations where RNA polymerase initiates and ends transcription; it can include more than one gene. Transcription unit refers to the DNA region that participates in transcription. A promoter, a structural gene, and a terminator are the three parts of the gene. A promoter is a DNA region that kicks off gene transcription. Promoters are found on the same strand and upstream on the DNA near the transcription start sites of genes. A structural gene, unlike a regulatory factor, codes for an RNA or protein product (i.e., regulatory protein). It could encode for a non-regulatory structural protein, enzyme, or RNA molecule.
Eventually, termination will result after the enzyme RNA polymerase bind to the DNA sequence know as terminator.
Types of RNA:
Ribosomal RNA (rRNA)
This is the most common kind of RNA found in cells.
Transfer RNA (tRNA)
The second most common form of RNA.
Messenger RNA (mRNA)
These are the forms of RNA that carry information to the ribosome about the amino acid sequence of a protein. Only messenger RNA is translated.
Heterogeneous nuclear RNA (hnRNA or pre-mRNA)
It is almost exclusively found in the nucleus of eukaryotic cells. It represents mRNA precursors, which are produced during the posttranscriptional processing of the molecule.
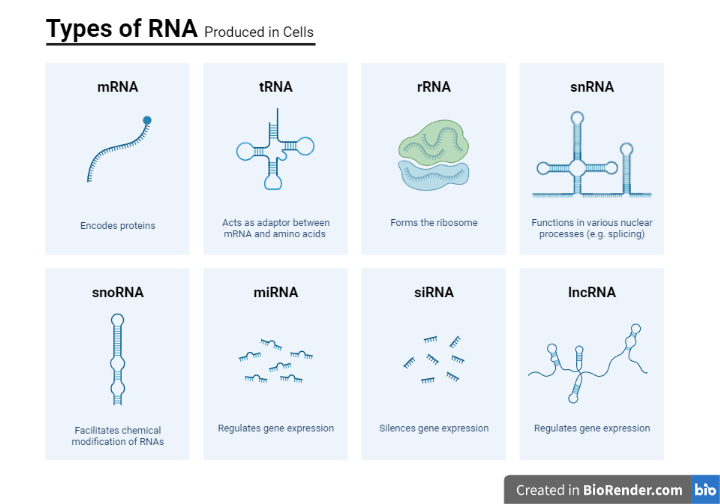
Small nuclear RNA (snRNA)
This type of RNA plays a major role during splicing (removal of introns) which is only found in the nucleus of eukaryotes.
Micro-RNA (miRNA)
A non-coding RNA with a length of 22 nucleotides that controls the expression or repression of other genes during development. Their main function is to regulate protein expression post-transcriptionally.
Ribozymes
These are the enzymatically active RNA molecules that are located in both prokaryotes and eukaryotes.
Components Required for DNA Transcription:
Transcription is carried out by an enzyme known as RNA polymerase, as well as a group of proteins known as transcription factors. Enhancer and promoter sequences are DNA sequences that transcription factors can bind to recruit RNA polymerase to a specific transcription site. The transcription initiation complex is made up of transcription factors and RNA polymerase operating together. The RNA polymerase starts mRNA synthesis by pairing complementary nucleotides to the original DNA strand, and this complex starts transcription. The mRNA molecule is elongated, and transcription is stopped once the strand has been fully produced. During the translation process, the freshly produced gene mRNA copies serve as templates for protein synthesis.
RNA polymerase:
The multisubunit enzyme RNA polymerase (RNAP) is the central enzyme of gene expression, responsible for transcribing a DNA sequence into an RNA sequence during transcription. The transcription process involves transcribing information from a molecule of DNA into a new molecule of messenger RNA. RNA polymerase is a complex molecule made up of protein subunits that regulates the process of transcription. All species have RNA polymerases; however, the number and content of these proteins varies by genus. Bacteria, for example, have only one type of RNA polymerase, but eukaryotes (multicellular creatures including yeasts) have multiple.
Eukaryotic RNA polymerase
RNA polymerase I
Most ribosomal RNA (rRNA) transcripts are synthesized by RNA polymerase I. These transcripts are made in the nucleolus, which is where ribosomes are assembled within the nucleus. Because these transcripts are directly associated with the synthesis of ribosomes, the availability of rRNA molecules synthesized by RNA polymerase can have an impact on important activities of cell biology.
RNA polymerase II
RNA polymerase II converts protein-coding genes into messenger RNA (mRNA). This 12-subunit enzyme operates as a complex that directly controls gene expression by synthesizing pre-mRNA transcripts. After being released by RNA polymerase II within the nucleus, biochemical changes prepare pre-mRNAs for translation. RNA polymerase II also produces microRNA (miRNA) molecules. These non-coding transcripts can mediate gene expression and mRNA activity after transcription.
RNA polymerase III
RNA polymerase III is responsible for converting rRNA genes into small RNAs such as transfer RNA (tRNA) and 5S rRNA that are essential for optimal cell function in both the nucleus and the cytoplasm.
RNA polymerase IV and V
RNA polymerase IV and V are transcription enzymes that originated from specialized versions of RNA polymerase II and are only present in plants. Both enzymes aid in the synthesis of small interfering RNA (siRNA) transcripts, which are involved in the silencing of plant genes.
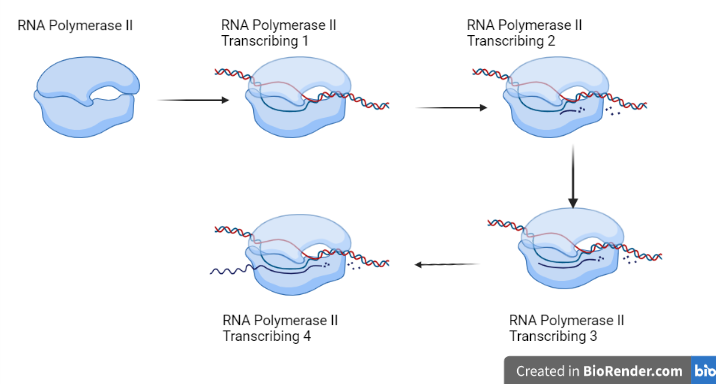
Fig: RNA polymerase II Transcribingcycle
Prokaryotic RNA polymerase
All of the genes in prokaryotes are transcribed by the same RNA polymerase. Polymerase in E. coli is comprised of five polypeptide subunits, two of which are identical. The holoenzyme is a polymerase that contains all five subunits.
The polymerase core enzyme is made up of four subunits, represented by the letters, α, α, β, and β. After a gene is transcribed, these subunits assemble, and when transcription is concluded, they disassemble. Each component serves a unique function.
- Two α-subunits are necessary to assemble the polymerase on the DNA
- β-subunit binds to the ribonucleoside triphosphate that will become part of the newly formed mRNA molecule
- β′ binds the DNA template strand.
Initiation of the transcription will only be proceeded with the involvement of this subunit. It confers transcriptional specificity, allowing the polymerase to commence mRNA synthesis from a specific initiation site. The core enzyme would transcribe from random sites if it didn’t have this unit.
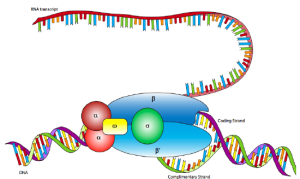
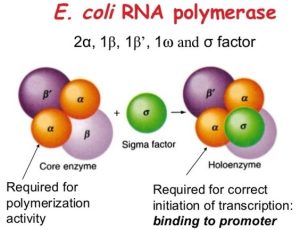
Fig: Structure of Prokaryotic RNA polymerase
Steps in Transcription:
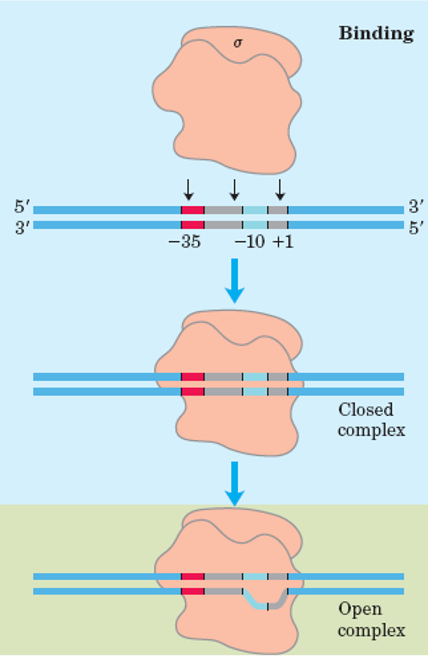
Initiation
The enzyme RNA polymerase catalyzes transcription by binding to and moving along the DNA molecule until it recognizes a promoter sequence. This region of DNA triggers the start of transcription, and a DNA molecule may have numerous promoter sequences. Transcription factors are proteins that regulate the rate of transcription and, similar RNA polymerase, bind to promoter regions.
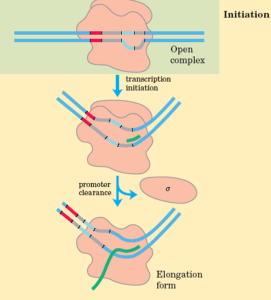
Elongation
Nucleotides are added to the mRNA strand. The unwound DNA strand is read by RNA polymerase, which then uses complementary base pairs to generate the mRNA molecule. The newly produced RNA is coupled to the unwinding DNA for a short period of time during this process. Adenine (A) from the DNA bonds to uracil (U) from the RNA during this procedure.
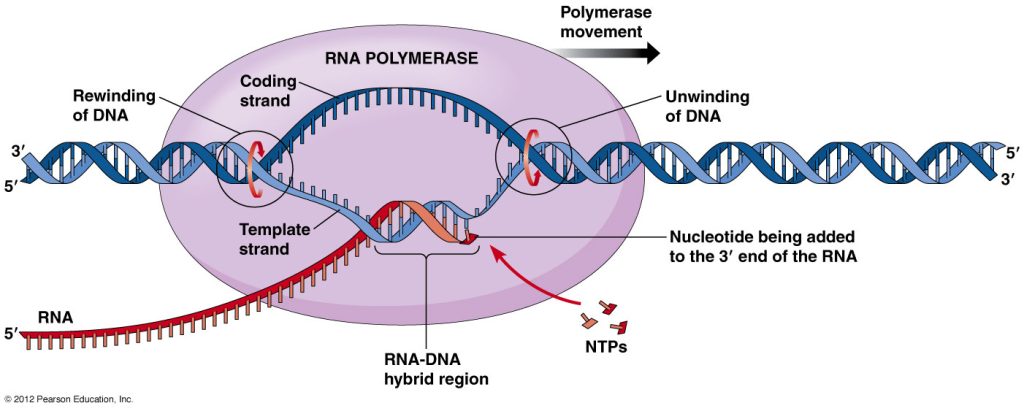
Termination
It occurs when RNA polymerase travels through a stop (termination) sequence in a gene, signaling the end of transcription. The mRNA strand is finished and separates from the DNA strand. Rho-dependent or Rho-independent termination are both possible.
Rho (ρ) dependent
The RNA molecule contains a binding site for a protein called the Rho factor, which binds to the DNA sequence during the termination process.
It begins to climb the transcript in the direction of the RNA polymerase and eventually reaches the transcription bubble. The Rho factor pulls the RNA transcript and the DNA template strand apart at the bubble, releasing the RNA molecule and stopping transcription. Later in the DNA, a transcription stop point sequence forces the RNA polymerase to halt, allowing the Rho factor to catch up and complete the termination process.
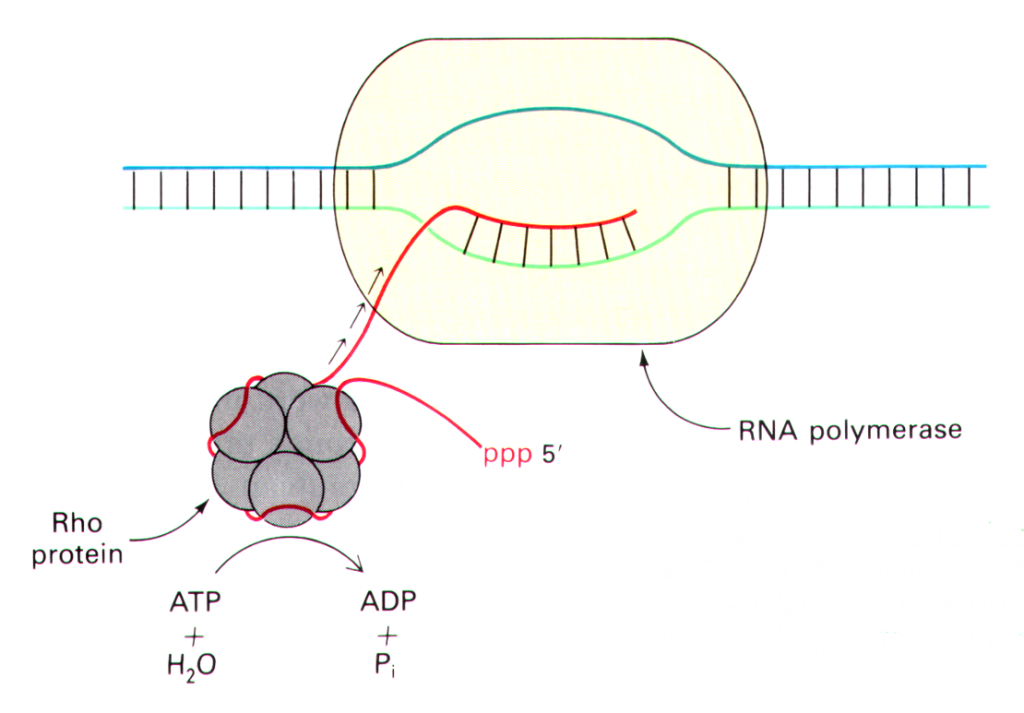
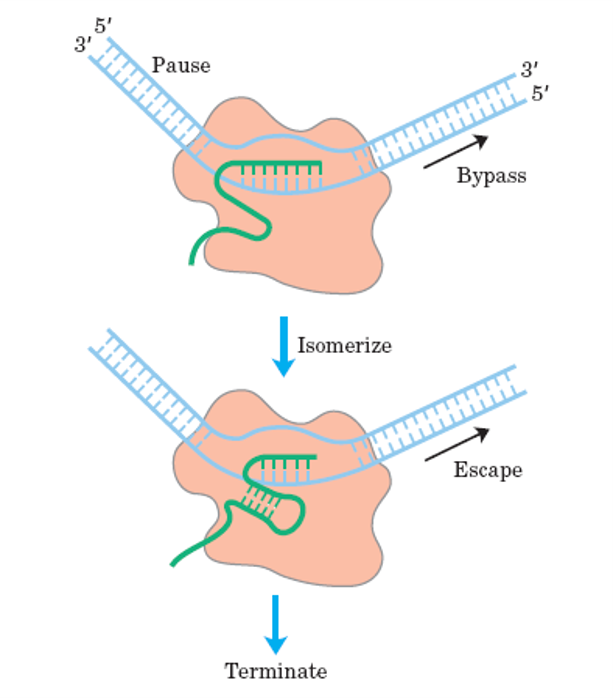
Fig: Types of termination of Transcription
Rho independent termination
The termination signal is a 30-40 nucleotide stretch on the RNA transcript that consists of a series of GC followed by a series of U. Because of the sequence specificity of this nascent RNA transcript, certain stem-loop structures will form to finish the transcription. The stem-loop structure causes RNA-pol to change conformation, causing it to stop traveling.
The interaction between the RNA-RNA hybrid and the DNA-DNA hybrid then lowers the stability of the DNA-RNA hybrid, causing the transcription complex to split.
Post transcriptional Modification:
Before translation can begin, RNA must be converted into a mature form. Post-transcriptional modification refers to the processing that occurs after an RNA molecule is transcribed but before it is translated into a protein. To control gene expression in the cell, the post-transcriptional process can also be regulated. No protein will be created unless the RNA is processed, translocated, and translated.
The three main post-transcriptional modifications are:
Capping
Capping occurs soon after transcription initiation, around the time the transcript reaches a length of 25–30 nucleotides before other modifications such as splicing of introns or addition of the poly(A) tail. The 5′ capping reaction replaces the triphosphate group at the 5′ end of the RNA chain with a special nucleotide that is referred to as the 5′ cap (Methyl guanosine tri phosphate).
Splicing
In eukaryotes, the structural gene has coding (exon) and non-coding (intron) regions. Removal of introns from the pre-mRNA, while linking the exons to form the mature mRNA is the main event that occur in splicing.
Tailing
At the end of transcription, 200-300 adenylate residues are added to the 3′ end of hnRNA to produce a poly-A tail. The poly(A) tail on mRNAs helps with transcription termination, mRNA export from the nucleus, and translation by protecting the mRNA molecule from enzymatic degradation in the cytoplasm.
RNA polymerase and drugs:
Because of its ubiquitous presence and function throughout life, RNA polymerase is a promising target for therapeutic development. Because of the biochemical distinctions in RNA polymerase between prokaryotes and eukaryotes, medicines that target microbial RNA polymerases.
Several antimicrobial medications work as RNA polymerase inhibitors by inhibiting the activity of bacterial or viral enzymes during a stage of transcription. The rifamycins5, for example, are a class of bacterial antibiotics that inhibit elongation by inhibiting the RNA polymerase exit channel. These antibiotics are widely used to treat difficult illnesses including leprosy and tuberculosis.
Drugs targeting the RdRp genes directly involved in viral genome replication and transcription are employed in the development of drug for SARS-CoV-2. Few analogs, such as Remdesivir (Ebola virus), Favipiravir (Influenza virus), NHC EIDD-1931 (wide spectrum), and Sofosbuvir, were originally developed to target RdRps of other RNA viruses (Hepatitis C virus).
Key points:
- In the process of gene expression, transcription is the initial step. It consists of making an RNA molecule by copying the DNA sequence of a gene.
- RNA polymerases, which join nucleotides to produce an RNA strand, are responsible for transcription (using a DNA strand as a template).
- Initiation, elongation, and termination are three phases of transcription.
- After transcription, RNA molecules must be spliced and have a 5′ cap and poly-A tail added to their ends in eukaryotes.
- Each gene in your genome has its transcription controlled separately.
References:
- Izban, M. G., & Luse, D. S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. Journal of Biological Chemistry 267, 13647–13655 (1992)
- Hirata A, Murakami KS. Archaeal RNA polymerase. Curr Opin Struct Biol. 2009 Dec;19(6):724-31. [PMC free article] [PubMed]
- Kritikou, E. Transcription elongation and termination: It ain’t over until the polymerase falls off. Nature Milestones in Gene Expression 8 (2005)
- Buonaguro, L., Tagliamonte, M., Tornesello, M.L. et al. SARS-CoV-2 RNA polymerase as target for antiviral therapy. J Transl Med 18, 185 (2020). https://doi.org/10.1186/s12967-020-02355-3
- Mooney RA, Landick R. RNA polymerase unveiled. Cell. 1999;98(6):687-690. doi:10.1016/s0092-8674(00)81483-x.