Introduction:
- In genetics, DNA fingerprinting is a method of extracting and identifying variable parts within the base-pair sequence of DNA (Deoxyribonucleic acid). It is also known as DNA typing, DNA profiling, genetic fingerprinting, genotyping, or identification testing).
- After noticing that certain sections of highly variable DNA (known as minisatellites), which do not contribute to the functioning of genes, are repeated inside genes, British geneticist Alec Jeffreys developed the approach in 1984.
- Jeffreys discovered that each human had a distinct pattern of microsatellites (the only exceptions being multiple individuals from a single zygote, such as identical twins).

Source: https://www.dreamstime.com/
- It is a technique for determining a living organism’s genetic makeup for determining the differences in satellite DNA regions in the genome. DNA fingerprinting is a laboratory technique used to establish a link between biological evidence and a suspect in a criminal investigation to determine identity or paternity.
- DNA can be extracted from relatively small amounts of biological evidence, such as a drop of blood or a single hair follicle.
- When DNA fingerprinting is performed and analyzed by expert forensic experts, the results can predict whether a person can be linked to a crime scene or be ruled out as a suspect quite accurately.
Principle:
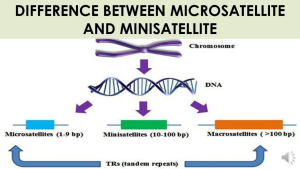
Minisatellite and microsatellite
99.9% of the human DNA on this planet is uniform. However, about 0.1 percent of DNA is unique in every individual. The human genome contains many small non-coding but inheritable base sequences that are repeated numerous times. They do not code for proteins but make-up 95 percent of our genetic DNA and therefore called the junk DNA. They are termed satellite DNA because they may be isolated from bulk DNA as satellites using density gradient centrifugation.
In satellite DNA, repetition of nucleotides is in tandem. Satellite DNAs are divided into microsatellites and mini-satellites based on their length, base composition, and number of tandemly repetitive units. A minisatellite is a 10-to-60-base-pair-long segment containing repeated DNA sequences. It normally occurs five to fifty times. The telomeres and centromeres of the chromosome are where these minisatellites are more noticeable. This short strand of repetitive DNA does not include any protein-coding sequences. Moreover, a microsatellite is also region of repetitive DNA that has a length of 1 to 6 (sometimes up to 10) base pairs known as short tandem repeats (STRs). It is also known as short tandem repeats (STRs). VNTR (variable number tandem repeats) refers to microsatellites as well as minisatellites.

Source: Google
At various loci on the chromosome the number of tandem repeats differs between individuals. There will be a fixed number of repeats for any specific location on the chromosome. Depending on the size of the repeat, the repeat areas are categorized into two kinds. Short tandem repeats (STRs) have 2-5 base pair repeats while variable number of tandem repeats (VNTRs) have repeats of 9-80 base pairs.
Because a child receives 50% of his DNA from his father and 50% from his mother, the number of VNTRs in a certain section of the child’s DNA will be varied due to insertion, deletion, or mutation in the base pairs. As a result, each individual has a unique VNTR composition, which is the basis for DNA fingerprinting.
Samples for DNA Fingerprinting Test:
DNA fingerprinting involves the purification of a person’s genome from a multitude of biological samples, including skin, hair, swab, skin, roots of the hair, saliva, blood, sweat, or other body fluids. However, blood is usually the easiest way.
Major Steps :
Stage 1: Sample Collection
DNA can be extracted from any biological sample or fluid. Buccal smear, saliva, blood, amniotic fluid, chorionic villi, skin, hair, body fluid, and other tissues are the most common types of samples used. In criminal scenarios, a buccal swab is routinely used. The procedure for obtaining samples with a buccal swab is straightforward and painless. If not properly maintained, it can easily get infected with microorganisms. Furthermore, a buccal swab’s DNA yield is extremely low. A blood sample can be used in place of the buccal swab sample. Another approach is to take a blood sample.
Stage 2: DNA Extraction
One of the most crucial stages in any genetic application is DNA extraction. Having good quality and quantity DNA improves your chances of getting positive results. Although any of the methods can be used to extract DNA, it is highly advised that you utilize a ready-to-use DNA extraction kit for DNA fingerprinting.
- DNA extraction using phenol-chloroform
- Extraction of DNA from CTAB
- DNA extraction with Proteinase K
Stage 3: Restriction Absorption, Enhancement or DNA Sequencing
Stage 4: Analysis of Results
Stage 5: Interpreting Results
Methods of DNA Fingerprinting:
Restriction fragment length polymorphism (RFLP)
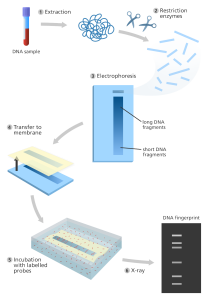
Restriction Fragment Length Polymorphism analysis was one of the earliest DNA analysis techniques discovered (RFLP). Alex Jeffreys discovered the methodology, which is based on an earlier Southern mapping procedure, in 1985. Restriction fragment length polymorphism (RFLP) is a method in which the genome is digested into small fragments using restriction (cleaving) enzymes.
A restriction fragment length polymorphism (RFLP) is a DNA sequence having restriction sites on both ends and a “target” sequence in the middle. For analysis, certain DNA locations (loci) are chosen, resulting in fragments of varying lengths between individuals. The DNA is cleaved into pieces at particular sequences using a restriction enzyme.
The generated DNA fragments are next separated by size using gel electrophoresis, which separates the fragments according to their molecular weight. The DNA is then transported to a DNA binding membrane once it has been sorted (blotting paper).
Then after, the membrane segments are incubated with a radioactive probe that will only bind to the target sequence, identifying its position on the DNA polymers.
The probe is identified by incubating it with an enzyme that turns it into a colour or light that may be seen on an x-ray film. Agarose electrophoresis is used to separate the fragments based on their size. We may use this technique to see if two samples originate from the same source or if they are independent. Because it was one of the earliest techniques for analysing DNA, RFLP technology has several drawbacks: it is slow and requires a big sample of DNA. However, because RFLP analysis is less susceptible to contamination than PCR, it is frequently utilized as a backup approach when contamination in the faster PCR analysis is suspected.
Source: https://www.yourgenome.org/
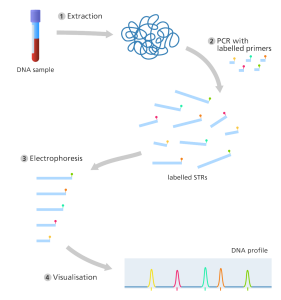
Polymerase Chain Reaction (PCR) amplification of short tandem repeats (STRs)
Polymerase chain reaction is the second most used method of DNA fingerprint analysis (PCR). This method is faster than RFLP and can be used with smaller DNA samples, however it is susceptible to contamination. In molecular biology, PCR is one of the most extensively utilized methods. Amplification of a specific segment of DNA by PCR is required for subsequent DNA fingerprinting methods. Within a test tube, PCR can produce millions of copies of a sample in just a few hours.
This enables more detailed testing on a limited amount of genetic material collected, as well as faster generation of a DNA profile. PCR, which is used to detect this, amplifies thousands of copies of a particular variable area. Gel-electrophoresis is used to amplify STR with a known repeat sequence and separate them. The STR’s migration distance is investigated.
A short synthetic DNA called primers is carefully engineered to connect to a highly conserved common non-variable section of DNA that flanks the variable region of the DNA for the amplification of STRs using PCR. The overall result of DNA fingerprinting will be determined by comparing the STR sequence size produced by PCR with other known samples.
Source: Genome Research Limited
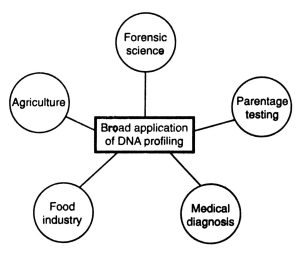
Application of DNA Profiling Technology:
- Investigations In Crime
- Molecular Archaeology
- Management of Wildlife
- Studies on organ transplantation.
- Forensic tests
- It’s possible to utilize it to establish paternity testing.
- To determine the frequency of specific genes in a population that causes diversity
- To utilize it to figure out how genetic drift has influenced evolution.
- To accurately establish paternity and/or maternity
Source: Google
References:
- Hypervariable ‘minisatellite’ regions in human DNA. Jeffreys AJ, Wilson V, Thein SL Nature 314: 67-73 (1985)
- Ramel C. (1997). Mini- and microsatellites. Environmental health perspectives, 105 Suppl 4(Suppl 4), 781–789. https://doi.org/10.1289/ehp.97105s4781
- Jeffreys, A J, V Wilson, SL Thein (1985b) “Individual-specific ‘fingerprints’ of human DNA.”
- Nature 316: 76-79.
- Gill P, Jeffreys AJ, Werrett DJ. Forensic application of DNA ‘fingerprints.’ Nature. 1985; 318:577–579.
- Weedn VW. Forensic DNA tests. Clin Lab Med. 1996; 16:187–196. [PubMed] [Google Scholar]

