Introduction To Bioinformatics:
- Bioinformatics is the application of computer technology to the management and analysis of biological data, such as genetic sequences and structures. It combines computer science, statistics, and molecular biology to understand and interpret biological data. It plays a crucial role in modern biology research, including genome sequencing and annotation, gene expression analysis, and protein structure prediction.
- Bioinformatics is essential for the analysis of high-throughput data generated by modern biological experiments, including genome sequencing, transcriptomics, and proteomics. It also plays a key role in the development of new drugs and therapies.
- Bioinformatics techniques and tools are used in a wide range of applications, including gene annotation, gene regulation, gene discovery, and drug development. It also aids in understanding the evolution of species, genetic variation, and the interactions between different genes and proteins.
- Some of the major fields in bioinformatics include sequence alignment and assembly, gene prediction, protein structure prediction, molecular evolution, and systems biology. The field is constantly evolving, with new technologies and computational methods being developed to analyse and interpret the large and diverse data sets generated by modern biology research.
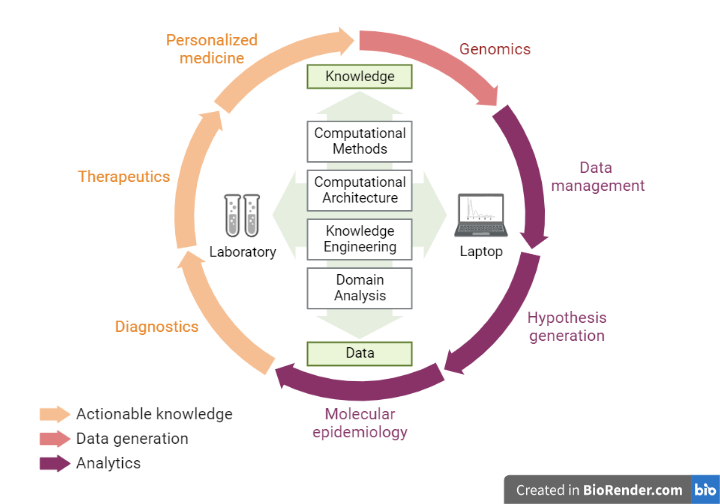
Fig: The Clinical Applications of Translational Bioinformatics
Biological And Bioinformatics Databases and Data Mining:
Biological databases and bioinformatics databases are collections of structured information that store, organize, and retrieve data related to biological systems and processes. These databases are designed to store, manage and integrate large amount of data from various sources. They are used to support the research in the field of bioinformatics, for example, to store and retrieve DNA and protein sequences, gene expression data, protein structures, and information about the interactions between different biomolecules.
Support Systems for Bioinformatics and Associated Scientific Disciplines:
Support systems for bioinformatics and associated scientific disciplines are a set of resources, tools and platforms that aid in the management, analysis, and interpretation of biological data. They are designed to help researchers, bioinformaticians, and scientists to access and use the vast amount of data and information that is generated by modern biological research. Some examples of support systems include:
Data repositories and databases
These are large collections of biological data that are publicly available, such as GenBank, UniProt, PDB, KEGG, DrugBank, and many others. They provide a centralized location for storing, managing, and sharing biological data, and they are often accompanied by web-based tools and interfaces for data retrieval and analysis.
Software and tools
These are the collections of programs and scripts that are used to perform various bioinformatics and computational biology tasks, such as sequence alignment, gene annotation, and phylogenetic analysis. Some popular bioinformatics software include BLAST, Clustal, T-Coffee, MUSCLE, EMBOSS, MAFFT, RNAfold, PSIPRED, PDBsum, Cytoscape, Bioconductor, Galaxy, KNIME, and many others.

Fig: Support Systems for Bioinformatics and Associated Scientific Disciplines
Workflows and pipelines
These are pre-defined sets of bioinformatics tools and commands that are used to perform specific tasks or analyses. They are designed to automate and streamline the data analysis process, and they are often accompanied by graphical user interfaces for easy use. Examples include the Broad Institute’s GATK (Genome Analysis Toolkit) and the EBI’s (European Bioinformatics Institute) Galaxy platform.
Cloud computing and High-Performance Computing (HPC) resources
Cloud computing and HPC resources provide access to powerful computing resources and storage, which are essential for bioinformatics and computational biology research, as they allow researchers to analyze and interpret large and complex data sets. Examples include Amazon Web Services (AWS), Google Cloud Platform (GCP), Microsoft Azure, and the Open Science Grid.
Training and support
Bioinformatics and computational biology require a multidisciplinary skill set, and many researchers may not have the necessary expertise. Therefore, training and support programs are available to help researchers learn the necessary skills. These include online tutorials, workshops, and graduate-level courses, and also bioinformatics support groups or core facilities within institutions.
Databases:
There are many databases in bioinformatics that store various types of biological information, such as DNA and protein sequences, genomic annotations, and functional information.
Some examples of commonly used biological and bioinformatics databases include:
GenBank and EMBL Large public databases that store DNA and RNA sequences from a wide range of organisms.
UniProt A database of protein sequences and functional information, including enzyme classifications, protein domains, and post-translational modifications.
PDB The Protein Data Bank, a repository of three-dimensional structures of proteins and other biomolecules.
KEGG A database of metabolic pathways and gene networks, linking genomic information to higher-order functional systems.
DrugBank A database of drugs, drug targets, and drug-target interactions.
Data mining:
It is the process of discovering patterns and insights from large sets of data. It involves the use of techniques from statistics, machine learning, and artificial intelligence to extract useful information from data
It is the process of extracting useful information from large datasets, and it plays an important role in bioinformatics. Data mining algorithms are used to identify patterns, relationships, and knowledge from the data stored in these databases. This can be used for tasks such as gene annotation, drug discovery, and the identification of biomarkers for disease.
In Bioinformatics, data mining methods are used to analyse large-scale data from high-throughput experiments such as microarray, RNA-seq, and proteomic data. These methods include clustering, classification, and association rule mining. Data mining techniques can be used to identify patterns in the data, such as the identification of differentially expressed genes in a cancer dataset or the discovery of new protein-protein interactions.
Sequence Analysis and Computational:
Sequence analysis is a branch of bioinformatics that involves the use of computational tools and methods to analyze DNA, RNA and protein sequences. The goal of sequence analysis is to understand the structure, function, and evolution of biological sequences, and it plays a crucial role in many areas of modern biology research.
There are several different types of sequence analysis, each with its own specific set of methods and tools:
Sequence alignment This is the process of comparing two or more sequences to identify regions of similarity. Alignment algorithms such as BLAST, Clustal, MUSCLE and T-Coffee are used to align sequences, and the results are used to identify homologous sequences and infer evolutionary relationships.
Sequence assembly This is the process of combining short sequences (e.g. from a genome sequencing project) into longer contigs or scaffolds. Assembly algorithms such as SPAdes, ABySS, Trinity and SOAPdenovo are used to assemble sequences, and the results are used to build a more complete representation of the genome or transcriptome.
Sequence annotation This is the process of adding functional and structural information to a sequence, such as gene prediction and functional annotation of genomic regions. There are many annotation tools, such as MAKER, MAKER-P, BRAKER, AUGUSTUS, and MAFFT
Phylogenetics This is the study of evolutionary relationships among organisms. Phylogenetic analysis is used to infer evolutionary relationships and construct evolutionary trees (phylogenies) based on DNA, RNA or protein sequences. Tools such as RAxML, IQ-TREE, MrBayes and PhyML are commonly used for phylogenetic analysis.
Models For Biological Data:
Models are simplified representations of a biological system or process, and they play an important role in bioinformatics and computational biology. There are several types of models that are commonly used to represent and analyse biological data, including:
Statistical models These models are used to describe the relationships between different variables in a dataset, such as the relationship between gene expression and disease status. Examples include linear regression, logistic regression, and mixed-effects models.
Machine learning models These models are used to discover patterns and relationships in data, using techniques such as clustering, classification, and regression. Examples include random forests, support vector machines, and neural networks.
Physical models These models are used to describe the physical properties and interactions of biomolecules, such as protein structures and ligand-protein interactions. Examples include molecular dynamics simulations, Monte Carlo simulations, and continuum models.
Network models These models are used to represent the interactions between different biomolecules, such as protein-protein interactions or gene regulatory networks. Examples include Bayesian networks, graphical models, and Boolean networks.
Biological models These models are used to represent the underlying biology of a system, such as metabolic pathways or gene regulatory networks. Examples include biochemical reaction networks, systems of differential equations, and Petri nets.
Biological Sequence Compression:
Biological sequence compression is the process of reducing the size of a biological sequence while maintaining the important information it contains. Compressing a sequence can make it easier to store, transmit, and analyse, and it also helps to reduce the time and computational resources required for sequence analysis.
There are several different techniques for compressing biological sequences, including:
Lossless compression This type of compression reduces the size of a sequence without losing any information. It is based on the redundancy and patterns present in the sequence, and it uses algorithms such as the Lempel-Ziv-Welch (LZW) algorithm, the Burrows-Wheeler Transform (BWT), and the Huffman coding.
Lossy compression This type of compression reduces the size of a sequence by removing some of the information it contains. It is based on the idea that some of the information in a sequence is not important for a specific analysis or application. Lossy compression methods are used when the information lost is not critical to the analysis or when the size of the data is a major concern.
Symbol coding This is a lossless compression technique that uses a variable-length code to represent each symbol in the sequence. For example, in DNA sequence, instead of using 2 bits for each nucleotide, A, C, G, T, it can use 2 bits for the most frequent nucleotides, A and T and 3 bits for less frequent G and C.
Run length encoding This is a lossless compression technique that replaces consecutive runs of identical symbols in a sequence with a single symbol and a count.
Reference-based compression This is a lossless compression technique that uses a reference genome to compress a sequence. It works by aligning the sequence to the reference genome and storing only the differences between the two.
Tools, software & Programmes:
Bioinformatics tools, software, and programs are used to analyse and interpret the large and diverse data sets generated by modern biology research. Some examples of commonly used bioinformatics tools include:
BLAST (Basic Local Alignment Search Tool) A widely used tool for comparing a DNA or protein sequence to a database of known sequences, to identify similarities and potential homologs.
Clustal A multiple sequence alignment tool, used to align multiple DNA or protein sequences.
T-Coffee A tool for multiple sequence alignment
MUSCLE A tool for multiple sequence alignment
EMBOSS A comprehensive collection of bioinformatics tools for sequence analysis
MAFFT A tool for multiple sequence alignment
RNAfol A tool for predicting the secondary structure of RNA molecules.
PSIPRED A tool for predicting protein secondary structure.
PDBsum A tool for visualizing and annotating the 3D structure of proteins and other biomolecules.
Cytoscape A tool for visualizing and analyzing complex networks, often used for protein-protein interaction networks.
There are also a number of bioinformatics software packages and platforms that include a variety of tools for specific tasks or for integrated data analysis. Some examples include:
Bioconductor An open-source software framework for bioinformatics, focused on the analysis of high-throughput data from genomics and proteomics experiments.
Galaxy An open-source, web-based platform for bioinformatics data analysis that offers a user-friendly interface and a large collection of tools.
R A programming language and software environment for statistical computing and graphics, widely used in bioinformatics for data analysis and visualization.
Python A programming language, often used in bioinformatics, with many libraries and frameworks such as Biopython, and Scikit-bio.
KNIME A free and open-source platform for data integration, analysis, and visualization, often used in bioinformatics.
References:
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J Mol Biol, 215(3):403–410.
- McGinnis, S. and Madden, T. L. (2004). BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res, 32(Web Server issue): W20–W25.
- Baxevanis, A.D., Bader, G.D. and Wishart, D.S. eds., 2020. Bioinformatics. John Wiley & Sons.
- Zvelebil, M.J. and Baum, J.O., 2007. Understanding bioinformatics. Garland Science.
