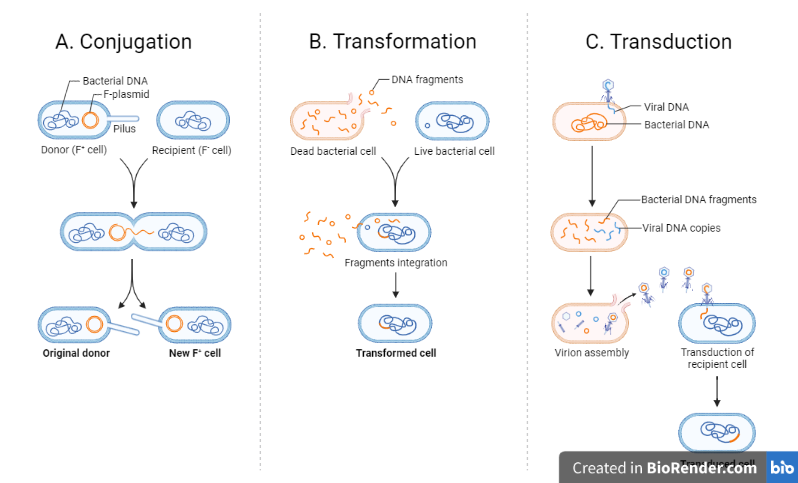
Bacterial recombination is the exchange of genetic material between bacterial cells that can occur via a number of ways. It is the process of genetic exchange between bacterial cells that results in the genetic variety and adaptation of bacterial populations to changing environments. It is characterized by the transfer of DNA from one species known as the donor to another known as the recipient.
Horizontal and vertical gene transfer:
Horizontal gene transfer (HGT) and vertical gene transfer (VGT) are two methods for transferring genetic material between organisms.
The transfer of genetic material from parent to offspring via reproduction is referred to as vertical gene transfer. This is the principal mechanism of gene transfer in most multicellular animals and certain asexually reproducing microorganisms. VGT, on the other hand, is responsible for organisms’ slow and steady evolution over many generations, as genetic features are passed down from parents to children. VGT, on the other hand, can result in the accumulation of harmful mutations over time, as well as the loss of desirable genetic variety due to genetic drift.
Horizontal gene transfer, on the other hand, refers to the exchange of genetic material between unrelated individuals. This can happen through bacterial mechanisms including conjugation, transformation, and transduction, as well as viral infection in eukaryotic cells. By facilitating the spread of genes that provide a selection advantage, such as antibiotic resistance or virulence traits, HGT can contribute to the rapid development of bacterial populations. It can also result in the transfer of genes between species, resulting in the production of new hybrid animals.
Mechanism:
There are several mechanisms of bacterial recombination, including transformation, transduction, and conjugation.
Conjugation
Bacterial conjugation is the transfer of genetic material, typically in the form of plasmids, from one bacterium to another via a pilus, a tiny protrusion that links two cells. This pathway is critical for the transmission of antibiotic resistance genes and virulence factors within bacterial populations.
The donor bacteria generate a pilus that extends to the recipient bacterium during conjugation, and a pore forms between the two cells. The plasmid DNA is subsequently transferred from the donor bacteria to the receiving bacterium via the pore. The plasmid DNA can be copied and expressed once inside the recipient cell.
Conjugation is a form of horizontal gene transfer in which bacteria share genetic material across individuals who are not genetically related. As a result, genes conferring beneficial features can swiftly spread through the community, resulting in rapid evolution of bacterial populations.

Fig: Bacterial conjugation
The general process of bacterial conjugation can be described as follows:
- The donor bacterium (also known as the male) produces a sex pilus, a thin, hair-like structure that extends out from the bacterial cell.
- The sex pilus attaches to a receptor site on the recipient bacterium (also known as the female), and the two bacteria are pulled closer together.
- A protein channel is formed between the two bacteria, which allows the transfer of genetic material from the donor cell to the recipient cell.
- The donor cell then sends a single-stranded copy of its plasmid DNA through the protein channel and into the recipient cell.
- Once inside the recipient cell, the plasmid DNA is copied to create a double-stranded molecule that can be integrated into the recipient’s genome, or remain as a separate plasmid.
- The recipient cell now has the genetic material from the donor cell, and can express any new genes it contains, including those that might confer new traits such as antibiotic resistance.
- The process of conjugation can continue as the recipient cell can also become a donor, producing its own sex pilus and transferring the genetic material to other bacteria.
Transduction
Transduction is a method of genetic transfer between bacteria in which bacteriophages (viruses that infect bacteria) are used as a vehicle for DNA transfer. A bacteriophage infects a bacterial cell and replicates within it, creating many copies of itself as well as the bacterial DNA it has acquired.
Transduction can cause the transfer of bacterial DNA across different species or strains of bacteria, resulting in the fast spread of genes conferring desirable features such as antibiotic resistance. Transduction can also take the form of specialized transduction, in which the bacteriophage carries a specific section of the donor DNA, usually from an integrated prophage in the donor bacterial genome.
The basic steps of transduction can be described as follows:
- A bacteriophage infects a donor bacterium and begins to replicate inside it.
- During the replication process, some of the bacterial DNA may be accidentally packaged into the viral particles (known as a bacteriophage), along with the viral genome.
- The bacteriophage containing the bacterial DNA is then released when the host cell lyses (bursts open), releasing the viral particles into the environment.
- The bacteriophage then infects a recipient bacterium, injecting its genetic material, which can include the bacterial DNA carried by the phage.
- The bacterial DNA from the donor bacterium may be incorporated into the genome of the recipient bacterium by recombination with the recipient’s own DNA.

Fig: Bacterial Transduction
There are two main types of transductions: generalized and specialized.
Generalized transduction
When a bacteriophage unexpectedly integrates bacterial DNA from the donor cell into the viral particle during the production of new virions, this is known as generalized transduction. As a result, some of the new phages generated from the donor cell have bacterial DNA rather than viral DNA. These phages can then infect a new bacterial cell, and the bacterial DNA can be transferred into the recipient cell during the viral replication process.
Specialized transduction
It happens when the bacteriophage that infects the donor cell integrates its viral DNA as a prophage into the bacterial chromosome. When the prophage is removed from the chromosome to begin the lytic cycle of viral replication, it may bring a tiny piece of neighboring bacterial DNA with it. As a result, certain genes are transferred from the donor cell to the receiving cell.
Transformation
Bacterial transformation is the process by which bacterial cells take up foreign genetic material from their environment, such as plasmids or DNA fragments, and incorporate it into their own genome. Frederick Griffith identified this mechanism in the bacteria Streptococcus pneumoniae in 1928. This procedure is significant in genetic engineering because it allows scientists to introduce new genes into bacterial cells for a variety of goals, including generating proteins of interest, investigating gene function, and discovering new medicines.
The steps involved in bacterial transformation are as follows:
- Preparation of bacterial cells: The preparation of the bacterial cells that will receive the foreign DNA is the initial stage in bacterial transformation. Bacterial cells grow in a nutrient-rich medium until they reach the exponential growth phase.
- Preparation of DNA: The foreign DNA that will be injected into the bacterial cells is isolated from a donor organism, either chemically or by specialist procedures like polymerase chain reaction (PCR). Following that, the DNA is purified to remove any impurities that may interfere with the transformation process.
- Introduction of DNA: Electroporation, heat shock, and chemical transformation are all methods for introducing DNA into bacterial cells. A brief electrical pulse is administered to the bacterial cells during electroporation, generating microscopic holes in their membranes through which foreign DNA can enter. Heat shock is quickly exposing bacterial cells to high temperatures, which destabilizes their membranes and allows DNA to enter. Chemical transformation involves treating bacterial cells with chemicals that make their membranes more permeable to DNA.
- Integration of DNA: After entering the bacterial cells, the foreign DNA may integrate into the bacterial chromosome or stay as a distinct plasmid. If the DNA integrates into the chromosome, it may recombine with the cell’s existing DNA, thereby changing its genetic composition. Instead, the DNA might be retained as a plasmid and replicate independently of the bacterial chromosome.
- Selection of transformed cells: A selectable marker is introduced with the DNA to identify the bacterial cells that have taken up the foreign DNA. This marker confers a property, such as antibiotic resistance, that distinguishes transformed cells from non-transformed cells. The converted cells can then be separated and cultured to generate new cells with the foreign DNA.
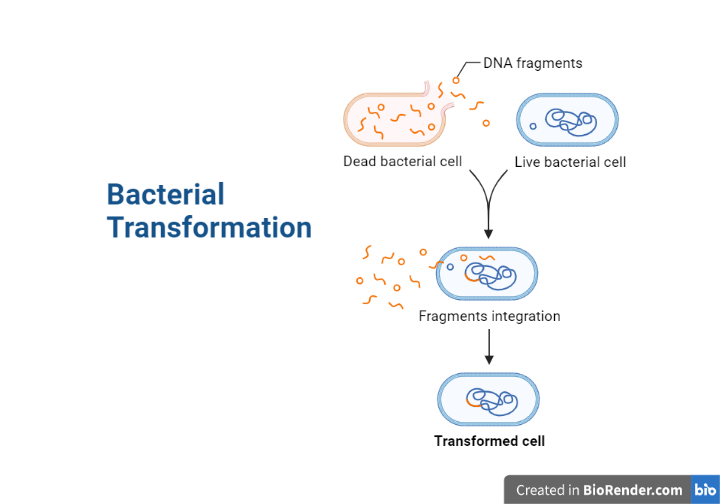
Fig: Bacterial Transformation
Impact of recombination on bacterial evolution:
The exchange of genetic material between two or more organisms, known as recombination, is a major driver of bacterial evolution. Bacteria can acquire new genes and features through recombination, which can give benefits such as antibiotic resistance, metabolic flexibility, and the ability to colonize new settings. Following are some examples of how recombination affects bacterial evolution:
Horizontal gene transfer: Recombination allows bacteria to acquire additional genes from other bacteria or even non-bacterial species horizontally. This can happen via mechanisms like transformation, conjugation, and transduction. Horizontal gene transfer can help favorable features spread throughout bacterial populations, such as antibiotic resistance, virulence factors, and metabolic pathways.
Hybridization: By recombination, bacteria can hybridize with other closely related species, resulting in the development of new bacterial lineages. This mechanism can result in the development of new bacterial species and the evolution of unique characteristics.
Genetic diversity: By recombining genetic material from multiple origins, recombination creates genetic diversity within bacterial populations. Its diversity can improve bacteria’s ability to adapt to changing surroundings and compete with other organisms.
Genome plasticity: Bacteria’s ability to recombine genetic material allows them to swiftly change their genomes in response to changing selection pressures. Its versatility enables bacteria to evolve new features and adapt to novel surroundings more quickly than mutation alone.
Current Methods for Recombination Detection in Bacteria:
The choice of method depends on the research question and the available data. A combination of methods is often used to increase the sensitivity and specificity of recombination detection in bacterial genomes. Detecting recombination events is essential for understanding bacterial evolution and the mechanisms driving the spread of bacterial pathogens. Here are some methods for detecting recombination in bacteria:
- Phylogenetic methods: Phylogenetic methods are based on the comparison of the evolutionary history of different genes or genomic regions. They involve the construction of phylogenetic trees based on the nucleotide sequences of the genes or regions of interest. Incongruence between these trees can be used as evidence for recombination events. Some popular methods in this category include the Neighbor-Joining method, the Maximum Likelihood method, and the Bayesian method.
- Statistical methods: Statistical methods rely on the comparison of the rates and patterns of evolution among different genomic regions to detect recombination events. For example, the Chi-squared test can be used to compare the distribution of nucleotide substitutions between different regions of the genome. The likelihood ratio test can be used to compare the fit of different models to the data, with one model allowing for recombination events and the other not. Other methods include the maximum-likelihood estimation (MLE) and the Bayesian estimation of recombination rates.
- Phylogenetic network analysis: Phylogenetic network analysis is a powerful method for detecting recombination events in bacterial populations. It involves the construction of a network of reticulations that connect different branches of the phylogenetic tree, representing the genetic exchange events that have occurred. This approach is particularly useful for detecting complex recombination patterns that cannot be detected by traditional phylogenetic methods.
- Comparative genomics: Comparative genomics involves the comparison of whole-genome sequences of different bacterial strains or species to identify recombination events. This approach can be used to identify genomic regions that are highly conserved within a bacterial species but divergent between species, indicating that they have been acquired through horizontal gene transfer.
- Recombination detection software: Several software tools have been developed for detecting recombination events in bacterial genomes, including RDP (Recombination Detection Program), GENECONV, Bootscan, MaxChi, and Chimaera. These programs use a combination of phylogenetic and statistical methods to detect recombination events and estimate their parameters.
References:
- Shikov AE, Malovichko YV, Nizhnikov AA, Antonets KS. Current Methods for Recombination Detection in Bacteria. Int J Mol Sci. 2022 Jun 2;23(11):6257.
- David M. Bonner, Genetic recombination in bacteria, Experimental Cell Research, Volume 9, Supplement, 1963, Pages 111-119.
- Didelot, X., & Maiden, M. C. (2010). Impact of recombination on bacterial evolution. Trends in microbiology, 18(7), 315-322.
- Fraser, C., Hanage, W. P., & Spratt, B. G. (2007). Recombination and the nature of bacterial speciation. Science, 315(5811), 476-480.
- Sánchez-Busó, L., Comas, I., & Jombart, T. (2019). Detection of recombination events in bacterial genomes from large population samples. Nucleic acids research, 47(4), e23-e23.