Telomere:
Introduction
- A telomere, a non-coding DNA element, is a specialized nucleoprotein structure located at the ends of linear eukaryotic chromosomes that acts as a protective “cap” to maintain genomic stability. Their length is about 800–1500 nucleotides long. Often compared to the plastic tips on shoelaces, telomeres prevent chromosomes from fraying, tangling, or fusing with one another.
- In prokaryotes, the DNA present is circular in the form of a circle, just like in plasmids. So, in prokaryotes there is an absence of telomeres.
- But in eukaryotes, because there are definite nuclei, a nucleus contains chromosomes, and the DNA present in those chromosomes is linear. So, just like telomeres in eukaryotes, they are present at the end portion of the telomere.
- The end portion is not equal in size. One strand is longer than the other strand. Now, let us look closely at the other strand. This is basically lagging strand. The other strand is leading strand.
- And the nucleotides present in telomeres are repetitive in nature. So, they have a sequence AATCCC in the leading strand. The complementary strand nucleotide is TTAGGG, in the lagging strand.
- This particular sequence is repeated again and again. So, the whole telomere region. The sequence is called non-coding hexameric sequence.
- The portion of the lagging strand of the single-strand DNA, this portion of DNA is coiled upon itself and makes a tiny knot-like structure called a T-loop (in order to protect from nucleases). And the single-stranded portion is tucked in a tiny portion, it is called D-loop or displacement loop.
- This loop prevents the telomere ends from being recognized. IT protects the 3′-end from being identified as damaged DNA, thereby preventing the activation of the ataxia-telangiectasia mutation and Rad3-related (ATM/ATR) damage response pathways.
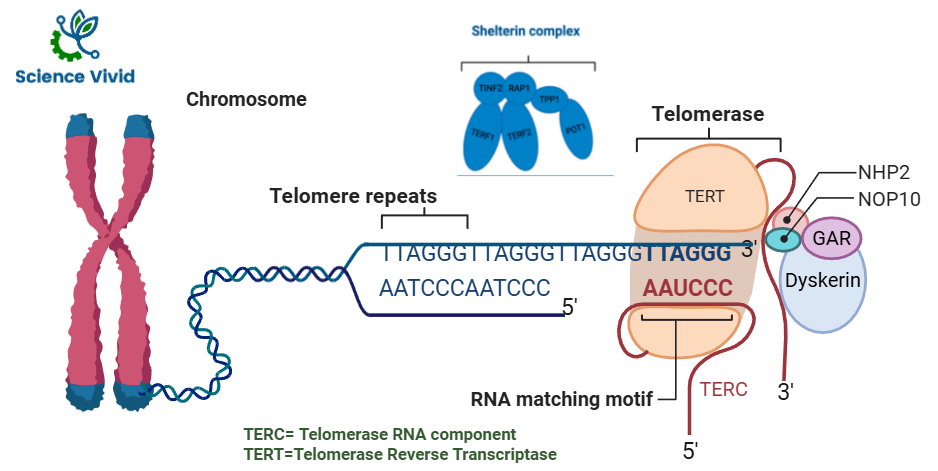
Fig: Structure of telomerase and Shelterin Complex
Structure and Composition
- DNA Sequence: In humans and all vertebrates, telomeres consist of tandemly repeated hexanucleotide sequences, specifically TTAGGG, repeated thousands of times (roughly 5,000 to 15,000 base pairs at birth).
- Shelterin Complex: A specialized group of six proteins (TRF1, TRF2, POT1, TIN2, TPP1, and RAP1) binds to the telomeric DNA to form a protective shield.
- Physical Shape: The 3′ end of the DNA often loops back and invades the double-stranded region, forming a T-loop and a D-loop (displacement loop) that effectively hides the chromosome’s end from being mistaken as a broken DNA strand.
The Shelterin Complex:
- The shelterin complex is a unique nucleoprotein assembly consisting of six proteins (TRF1, TRF2, Rap1, TIN2, TPP1, and POT1) that binds exclusively to telomeric DNA. It is necessary for telomere protection, telomere signaling, length management, and genome integrity. It is also known as telosome.
- The shelterin complex remain attached to the T-loop and maintain the structure.
- Shelterin prevents abnormal DNA damage response (DDR) by differentiating between DNA double-strand breaks and native chromosomal ends.
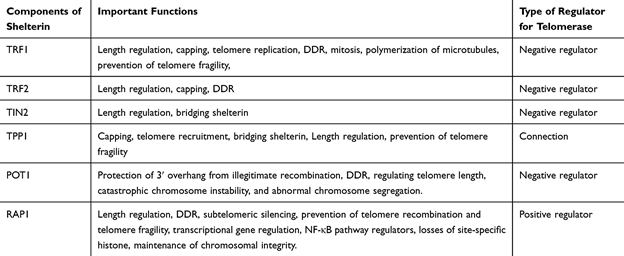
Fig: The Role of the Components of Shelterin Complex in Telomeres and Telomerase Regulation
Source: Clinical Interventions in Aging 2020:15
What is the Hayflick Limit?
- Leonard Hayflick (1928–2024), the American anatomist discovered theory of limitation of normal cell division.
- The Hayflick limit (also called the Hayflick’s phenomenon) is defined as the finite number of times a normal human somatic (body) cell population can divide before it permanently stops dividing and enters a state of cellular senescence phases.
- In typical experiments with human fibroblasts (skin cells), this limit is around 40–60 divisions
Hayflick outlines the three stages that typical cultivated cells go through.
- Hayflick outlines the three stages that typical cultivated cells go through. • He called the primary culture “phase one” when he first started his experiment.
- The second phase is characterized by cell proliferation, which Hayflick dubbed the “luxuriant growth” period.
- Following months of cell division, the cells finally enter phase three, a stage he dubbed “senescence,” in which the rate of cell division decreases and eventually stops.
Telomerases:
- Telomerase, the immortality enzyme, is ubiquitous in all mammalian embryonic tissue and remains active in germs cells but is down-regulated in most somatic tissues.
Telomerase is an enzyme with two subunits: TERT and TERC
1.TERT (Telomerase reverse transcriptase) is basically the protein part of this enzyme and functions as a reverse transcriptase. It is the catalytic subunit responsible for enzyme activity in telomerase, is the rate-limiting factor of human telomerase enzyme activity. Two of the critical telomere-specific proteins involved in the regulation and maintenance of the telomere length are the shelterin and CST complexes. A CST complex (CTC1-STN1-TEN1), a heterotrimeric protein complex, that binds single-stranded (ss) DNA that help in DNA replication and also protect telomeres.
2. TERC (Telomerase RNA component) is an RNA template, and it attaches more nucleotides to the telomere to complete its length (synthesis of telomeric repeat DNA).
- In eukaryotic chromosome, telomeres maintain the integrity of the genome by preventing chromosomes fusing, chromosomal rearrangement, and also maintain complete replication of the linear DNA molecules.
- A short region at the 3′ end will remain single-stranded, and this is what telomerase will act on and will result in the addition of many repeat sequences, from a few dozen to a few hundred, which prevents chromosome end shortening.
- The DNA polymerase further functions after telomerase action. DNA polymerase synthesizes a DNA strand complementary to the RNA primer found on the telomerase and subsequently translocate to the end of the newly synthesized strand, and the process repeats itself.
- Many cycles of repeats can occur. Once telomerase has completed its function, DNA primase synthesizes an RNA primer near the 3′ end, and DNA polymerase fills in the vacant region.
Telomerases and cancer
- Since telomerase activity controls cellular proliferation, it needs to be strictly controlled to avoid causing carcinogenesis.
Cancers may arise from mutations in the genes encoding these complexes. Therefore, considering the therapeutic approaches to treat such diseases, it is essential to fully understand the molecular processes of these proteins. - The human hTERT gene, located at chromosome 5p15.33, encodes the catalytic protein subunit known as telomerase reverse transcriptase (TERT), which is the reverse transcriptase telomerase.
- Human telomerase RNA (hTR) or human telomerase RNA component (hTERC) is an important RNA component that is encoded by the hTERC gene located on chromosome region 3q26. • hTR is involved in the catalysis, localization, and assembly of the telomerase holoenzyme in addition to serving as a template (carrying sequence complementary to one or more copies of telomeric repeats) for the synthesis of telomere DNA.
- Telomerase is implicated in gene expression regulation, cell proliferation, apoptosis, WNT/β-catenin signaling, NF-kB signaling, MYC-driven oncogenesis, DDR, cell adhesion and migration, and epithelial–mesenchymal transition, according to recent studies.
- It is believed that each of these telomerase functions has a major role in the oncogenesis process.
