Introduction:
Crossing over is the refers to the interchange of genetic material between non-sister chromatids of homologous chromosomes involving breakage and reunion at precise point during meiotic prophase (pachytene), which results in new allelic combinations in the daughter cells. In 1912, the term crossing over was first used by Morgan and Cattell.
Diploid cells have two sets of chromosomes, one from each parent, known as homologous chromosomes. Diploid cells go through meiosis to create haploid gametes during sexual reproduction. During meiosis, in prophase I, homologous chromosomes align and exchange genetic material, resulting in recombinant chromosomes containing a mix of maternal and paternal genes.
Types:
Bases in the cell
Basically, there are two types of crossing over as per to its occurrence in the somatic or germs cells.
Somatic/mitotic crossing over
Somatic or mitotic crossing over occurs when the process of crossing over happens in the chromosomes of an organism’s somatic cells during mitotic cell division. It has no genetical significance and rarely occur.
Germinal or meiotic crossing over
When the process of crossing over occurs in germinal cells during the gametogenesis in which the meiotic cell division takes place, it is known as germinal or meiotic crossing over. It is universal and its occurrence and is of great genetic significance.
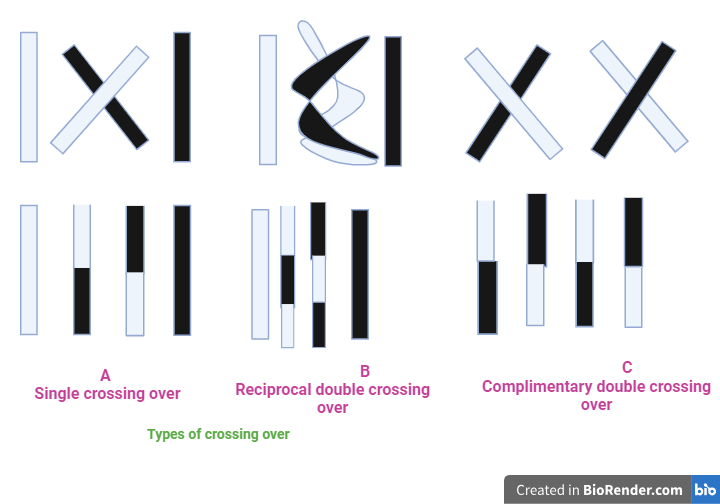
Fig: Types of crossing over
Based on the number of chiasmas
According to the number of chiasmas following types of crossing over:
Single crossing over
When the chiasma occurs only at one point of the chromosome pair when the crossing over is known as single crossing over. It produces two crosses over chromatids and two non-cross over chromatids.
Double crossing over
When the chiasmata occur at two pints in the same chromosome, the crossing over is know as double crossing over. Each chiasma forms independently of the others, and possibility of four types of recombination can occur. In the double crossing over, following two types of chiasmas may be formed.
Reciprocal chiasma:
In these types of crossing over, the same two chromatids are involved in the second chiasma formation as in the first chiasma. Thus, the second chiasma restores the order disrupted by the first chiasma and results in the formation of two non-cross over chromatids. In the reciprocal chiasma, out of four chromatids only two are involved in the double crossing over.
Complimentary Chiasma:
When both of the chromatids taking part in the second chiasma are different from those chromatids involved in the first chiasma, it is known as complimentary chiasma. It produces no non-crossover and four single cross overs. It occurs when three or four single chromatids of the tetrad undergo the crossing over.
Multiple crossing over
When crossing over takes place at more than two place in the same chromosome pair, then it is known as a multiple crossing over. It occurs rarely.
Factors controlling frequency of crossing over:
The frequency of crossing over is mostly determined by the distance between associated genes. However, a variety of genetic, environmental, and physiological factors influence the incidence of crossing over. Thy are as follows:
- Temperature: The frequency of crossing over is increases by high and low temperature increases.
- Radiation: The radiations increase the frequency of crossing over.is increases by the exposure to the x-ray and other.
- Age: The frequency of crossing over decreases with the increases in age.
- Distance: The frequency of crossing over increases with the increase in the distance between the genes.
- Chemicals: While some chemical increase t he frequency of crossing over, other chemicals decrease it.
- Inversions: Inversions supress the crossing over.
- Chiasmata formation: The formation of one chiasma discourages the formation of their chiasmata in the near vicinity.
- Centromere: Generally, genes present close to the centromere show reduced crossing over.
Chromosomal Maps:
Gene mapping is the assignment of a gene to a locus on a chromosome. By, mapping the relative positions of many genes and other markers it is possible to generate a chromosome map or a map of entire genome. Gene maps are of following types:
Genetics/linkage/chromosome map: It assigns genes to linkage groups.
Cytogenetic map: It is created by correlation phenotypes with observable chromosomal rearrangements and deficiencies.
Physical or molecular map: It is created by ordering cloned fragments of genomic DNA and dis calibrated in real units, i.e., bp, kbp, etc. The physical map has the highest resolution and is the ultimate aim of the mapping projects.
Chromosome Maps:
The method of construction of maps of different chromosomes is called genetic mapping or chromosomal mapping. It includes the following process:
Determination of linkage groups
Before starting the genetic mapping of the chromosome of a species, one has to know the exact number of chromosomes of the species and then determine the total number of genes of that species through hybridisation experiment. Moreover, it is also possible to determine different linkage groups of a species through the hybridisation technique. Linked group is formed by all the linked genes of a chromosome. The number of linkage groups of a species corresponds with haploid chromosome numbers of species.
Determination of map distance
The intergene distance on the chromosomes cannot be measured by units employed in light microscopy but map units. A map unit is equal to 1% of cross overs (recombination) for e.g. If a F1 hybrid having the genotypes Ab/aB produces 8% of cross over gametes AB and ab, then the distance between A and B is estimated at 16 maps units. As each chiasma produces 50% crossover products, it is equivalent to 50 map units. If the mean number of chiasmata is known for a chromosome pair, the total length of the map for the linkage group may be predicted as
Total length= mean number of chiasmata × 50
The % of crossing over between two linked genes is calculated by test crosses in which a F1 dihybrid is crossed with a double recessive parent e.g.
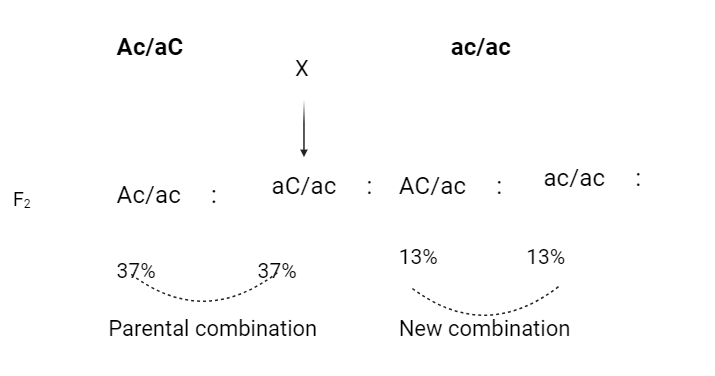
Fig: Determination of map distance
The last two groups were produced by crossover gametes (13+3) from the dihybrid parent. Thus, 26% of all gametes were of cross types and the distance between the loci A and C is estimated at 26 map units.
A three-point test cross or trihybrid test cross gives us information regarding relative between genes and also shows linear order in which these genes should be present on chromosome.
To find the distance between A and b count all single and double cross overs i.e., 18% +2%=20% or 20 map units distances between A and B. To find the distance between B and C, 8%+2%=10% or 10 maps units distance between B and C. Therefore, the distance between A and Cis 30 map units. When double crossovers are detected in a 3-point linkage experiment it is 26 map units.
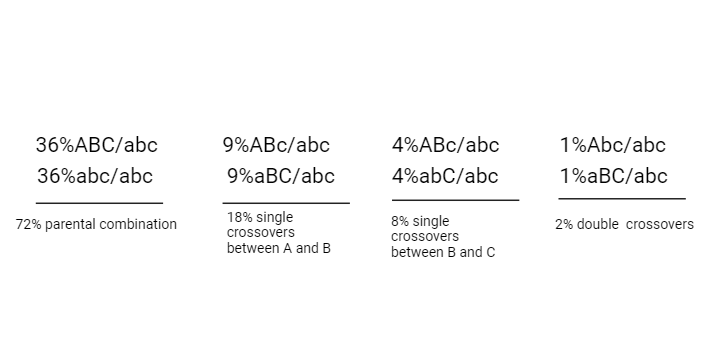
Fig: Determination of map distance
Determination of Gene order
After determining the relative distance between genes of a linkage group, it becomes easy to place genes in their proper linear order. For example, distance between the genes A-B=12, B-C=7 and A-C=5 and we can determine the order of genes in the following manner.
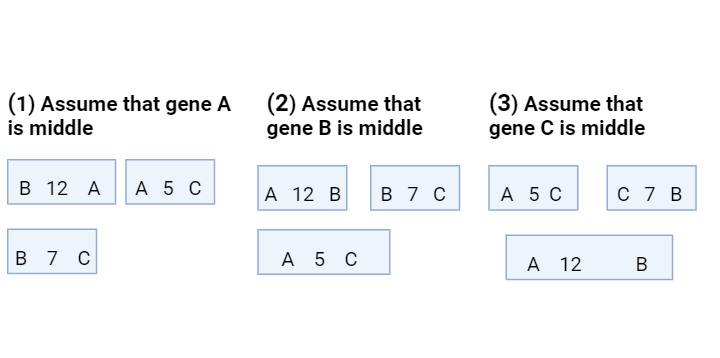
Fig: Determination of Gene order
In the third case, the distance between A-B is equitable, so gene C must be in middle.
- Combining map Segments: Finally, the difference segments of maps of a completer chromosome are combined to form a complete genetic map for a chromosome.
Significance of chromosome maps:
- Its shows the number of chromosomes of number of linkage group of organisms
- It shows exact location, arrangement and sequence of genes on chromosome.
Interference:
In 1911, Muller discovered the phenomenon of interference. Its higher organism one chiasma formation reduces the probability of another chiasma formation in an immediately adjacent region of the chromosome, probably because of physical inability of the chromatids to bend back upon themselves within certain minimum distance. This tendency of one crossover to interface with the other crossover is called interference. Interference is inversely proportional to the distance between the points of crossing over, i.e., interferences is maximum over short distance and decreases as the distance increases.
Types of interference:
Positive interference
When one crossover inhibits another crossover. Thus, when the double crossovers are absent, we can say that interference is cent per cent.
Negative interference
where one crossover stimulates another. Recombination between vary close markers shows evidence of negative interference, i.e., several crossovers appear to be clustered.
Interference varies from chromosome to chromosome and within the chromosome form one point to the other. Generally, it is maximum near the centromere and at the two ends of the chromosome (telomere).
Coincidence:
Coincidence or coefficient of coincidence is inverse measure of interference and is expressed as the ration between the actual number of double crossovers and the expected number of such double cross overs, i.e.

Fig: Coincidence
The coincidence is the complement of interference, o coincidence+ interference= 1.0
i.e., when interference is completer (1.0), no double crossovers will be observed and coincidence becomes zero. When interference decreases, coincidence increase. Coincidence is generally quite small for short map distance.
Significance of crossing over:
- The crossing over provides a source of genetic variation. The origin of new characters due to the exchange of a segment from one chromosome to another.
- The crossing over provides the direct evidence that the genes are linearly arranged on the chromosome
- Chromosome mapping is possible by the study the frequency of crossing over
- Crossing over is very important in breeding to improve the variety of plants and animals.
