Introduction:
Maxam-Gilbert sequencing is a DNA sequencing technology invented by Allan Maxam and Walter Gilbert in the late 1970s. It was one of the first ways of sequencing DNA, preceding the more frequently used Sanger method.
Maxam-Gilbert sequencing uses chemical treatment of DNA to induce precise breaks in the backbone of the molecule at known sites, followed by fragment separation and identification.
The Maxam-Gilbert method was created as one of the first DNA sequencing methods and was widely utilized in the early days of DNA sequencing. However, because of its relative difficulty and cost in comparison to more current sequencing methods such as the Sanger sequencing method and next-generation sequencing technologies, it is now less widely utilized.
Steps:
The Maxam-Gilbert method involves several steps:
Labelling of DNA
The first step involves labelling the DNA molecule with a radioactive or fluorescent label. This allows the fragments of the DNA to be visualized on an autoradiogram or gel.
Chemical cleavage
The labelled DNA is then treated with chemicals that cleave the DNA at specific positions. There are four different chemical treatments that are used in the Maxam-Gilbert method, each of which cleaves the DNA at different positions:
Hydrazine cleaves DNA at guanine (G) and adenine (A) residues.
Dimethyl sulfate (DMS) cleaves DNA at cytosine (C) and guanine (G) residues.
Formic acid cleaves DNA at purine residues (A and G).
Piperidine cleaves DNA at purine residues (A and G).
Fragment separation
After the DNA has been cleaved, the resulting fragments are separated by size using gel electrophoresis.
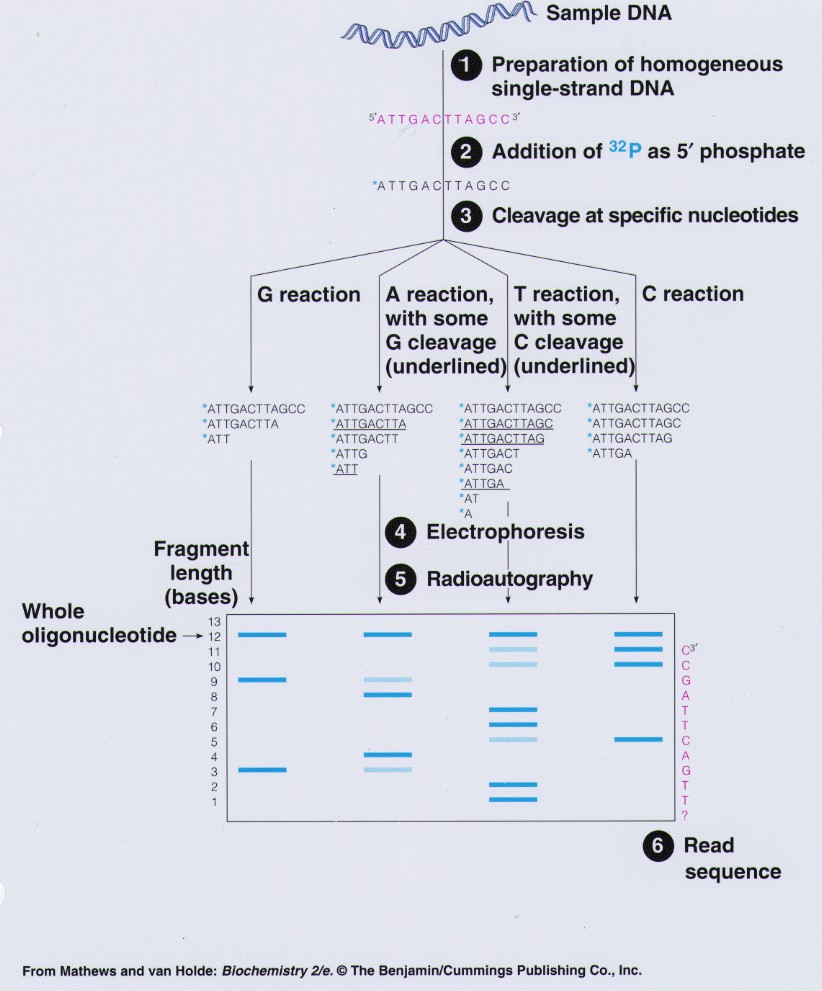
Fig: Steps of DNA sequencing by Maxam-Gilbert technology
Visualization and detection
The fragments are then visualized using autoradiography or other detection methods. The pattern of fragments generated by the chemical cleavage is then analysed to determine the sequence of the DNA.
Advantages:
- High accuracy: The Maxam-Gilbert method can provide high accuracy in determining the DNA sequence, as the chemical cleavage is precise and predictable.
- Can detect modifications: The Maxam-Gilbert method can detect modifications to the DNA, such as methylated or damaged bases.
- Applicable to different DNA types: The Maxam-Gilbert method can be used to sequence different types of DNA, including single-stranded DNA and RNA.
- Can sequence through difficult regions: The Maxam-Gilbert method can sequence through difficult regions of DNA, such as regions with high GC content or secondary structures.
Disadvantages:
The Maxam-Gilbert method, also known as the chemical cleavage method, was one of the earliest DNA sequencing methods but has several disadvantages compared to more modern sequencing technologies. Some of the disadvantages of the Maxam-Gilbert method include:
- Complexity and cost: The Maxam-Gilbert method is a relatively complex and costly method of DNA sequencing, which has limited its use in modern DNA sequencing technologies.
- Limited read length: The read length of the Maxam-Gilbert method is limited to around 50-500 nucleotides, depending on the specific conditions used. This is much shorter than the read lengths that can be achieved with modern sequencing technologies.
- Radioactive or fluorescent labelling: The Maxam-Gilbert method requires the use of radioactive or fluorescent labelling, which can be hazardous and require special handling.
- Time-consuming: The Maxam-Gilbert method is a time-consuming process, as it involves several steps including labelling, chemical cleavage, fragment separation, and detection.
- Inconsistent yields: The yields of cleaved fragments can be variable and difficult to predict, which can affect the accuracy of the sequencing results.
- Not suitable for high-throughput sequencing: The Maxam-Gilbert method is not suitable for high-throughput sequencing, as it is a manual and time-consuming process.
Applications:
The Maxam-Gilbert method was used extensively in the early days of DNA sequencing and played a crucial role in the Human Genome Project. Some of the applications of the Maxam-Gilbert method include:
- DNA sequencing: The Maxam-Gilbert method was one of the first techniques used to sequence DNA. It was used to sequence the genomes of many organisms, including bacteria, viruses, and humans.
- Mutational analysis: The Maxam-Gilbert method can be used to identify mutations in DNA. By comparing the sequence of a mutated DNA sample to a wild-type sample, the location and nature of the mutation can be determined.
- Mapping protein-DNA interactions: The Maxam-Gilbert method can be used to map the location of protein-DNA interactions. By selectively cleaving DNA at sites where proteins are bound, the binding sites can be identified and mapped.
- Structural analysis of DNA: The Maxam-Gilbert method can be used to study the structure of DNA. By selectively cleaving DNA at different sites, the location and nature of structural features such as bends and twists can be identified.
- Characterization of DNA modifications: The Maxam-Gilbert method can be used to identify and characterize DNA modifications such as methylation. By selectively cleaving modified DNA at specific sites, the presence and location of modifications can be determined.
References:
- Maxam, A. M., & Gilbert, W. (1977). A new method for sequencing DNA. Proceedings of the National Academy of Sciences, 74(2), 560-564.
- Hagenbuchle, O., & Wellauer, P. K. (1977). Chemical sequencing of DNA by the Maxam-Gilbert method. Methods in enzymology, 51, 346-359.
- Smith, L. M., & Hood, L. E. (1983). The identification of protein-DNA contact sites by DNA sequencing. Proceedings of the National Academy of Sciences, 80(14), 4394-4398.
- Sanger, F., Nicklen, S., & Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences, 74(12), 5463-5467.
- Zhou, X., & Wang, E. (2011). DNA sequencing by synthesis using pyrosequencing technology. Methods in molecular biology (Clifton, NJ), 733, 119-131.
- Ju, J., Ruan, C., Fuller, C. W., & Glazer, A. N. (1995). Methods for sequencing DNA by hybridization with oligonucleotide matrix. Genome research, 4(6), 537-547.
