Introduction:
The cytogenetic method known as fluorescence in situ hybridization (FISH) was developed in the early 1980s. FISH produces coloured signals that may be seen under a fluorescence microscope by utilizing fluorescent DNA probes to target particular chromosomal regions within the nucleus.
It is a macromolecule recognition approach that is regarded as a new development in the field of cytology for identifying specific DNA sequences, diagnosing genetic diseases, mapping genes, and discovering new oncogenes or genetic abnormalities causing various types of cancer within a cell or tissue sample.
FISH is a simple technique that involves hybridizing a DNA probe to its complementary sequence on chromosomal preparations that have been previously placed on slides. Probes are labeled either directly with fluorescent nucleotides or indirectly with reporter molecules that are then recognized by fluorescent antibodies or other affinity molecules. Finally, microscopy analysis is used to visualize probes and targets in situ.
The main advantage of this visually appealing technique as a combined molecular and cytological approach is its unique capacity to provide an intermediate degree of resolution between DNA analysis and chromosomal investigations, while also keeping information at the single-cell level.
Principle:
FISH works on the basis of a labeled probe specifically binding to complementary DNA or RNA sequences in a sample. The probe is tagged with a fluorescent dye and can be made of RNA or DNA. When the probe is incorporated into the sample, it binds to complementary sequences, either within the same chromosome or between chromosomes.
The probes are either directly labeled by incorporating a fluorophore or indirectly by incorporating a hapten. Following denaturation, the labeled probe and the target DNA are combined, allowing complementary DNA sequences to anneal. If the probe had been indirectly labeled, it would be necessary to perform an additional enzymatic or immunological detection step in order to see the non-fluorescent hapten. The signals are then examined using fluorescence microscopy. Fluorochrome, which emits colourful signals at the hybridization site, is a component of the enzymatic detection. The immunological detection approach is based on antibody binding to certain antigens, which is subsequently demonstrated by a coloured histochemical reaction observable under a light microscope or fluorochromes under UV light.
The specificity of the probe-target binding is critical for the effectiveness of FISH assay. The probe is designed to connect solely to specific sequences, which are frequently unique to a specific gene, chromosome, or cell type. Because of this specificity, FISH can be used to identify individual genes or chromosomes, visualize gene expression patterns, and detect chromosomal abnormalities.
Probes:
Probes are short single-stranded DNA or RNA molecules that have been fluorescently tagged. These probes are programmed to bind to specific DNA or RNA sequences in a sample. Because of the dye’s fluorescence, the location of the target sequence can be observed and studied. It has a property of hybridizing through complementary base pairing, resulting in the formation of double-stranded hybrids. The probes emit a fluorescence signal when they bind to their target, which can then be visualized under a microscope.
There are different types of probes that can be used in FISH. Probes can be designed to target specific sequences based on the genetic information available for the organism being studied. The probes can be made from either synthetic or naturally occurring DNA or RNA. In order to maximize specificity, probes are often designed to be complementary to the target sequence, but not to other regions of the genome.
Locus specific probes
These probes bind to a specific area of a chromosome. These probes can detect microdeletion and microduplication syndromes within a specific chromosome.
Whole-chromosome probes
These probes are collections of tiny probes tagged with distinct fluorescent dyes, each of which binds to a different sequence throughout a given chromosome. These probes can be used to map the entire chromosome, providing information on chromosomal abnormalities. Scientists can label each chromosome in its own distinct colour by using numerous probes labeled with a mixture of various fluorescent dyes which is known as a spectral karyotype that includes full-colour map of the chromosome.
Repetitive probes
The repeating sequences located in the centre of each chromosome are used to create repetitive probes, which can be used to examine whether a person has the appropriate number of chromosomes. These probes can be used in conjunction with locus-specific probes to identify whether a person lacks genetic material from a given chromosome.
In FISH, fluorescent probes are used to visualize specific DNA or RNA sequences in a sample. There are several types of fluorescent probes that can be used, including:
- Cy3: A green fluorescent dye that is commonly used in FISH experiments.
- Cy5: A red fluorescent dye that is also commonly used in FISH experiments.
- FITC (fluorescein isothiocyanate): A green fluorescent dye that is often used in FISH experiments.
- Texas Red: A red fluorescent dye that is commonly used in FISH experiments.
- Rhodamine: A red fluorescent dye that is sometimes used in FISH experiments.
- DAPI (4′,6-diamidino-2-phenylindole): A blue fluorescent dye that is often used to stain DNA in FISH experiments.
- Alexa Fluor dyes: A family of fluorescent dyes that are available in a wide range of colors, including green, red, and blue. Alexa Fluor dyes are commonly used in FISH experiments.
Steps:
The steps in Fluorescence In Situ Hybridization (FISH) technique typically include:
Sample preparation
Obtain a biological sample, such as tissue or cells, and prepare it for FISH by fixing and permeabilizing the cells. The sample (e.g., tissue section or cells) is fixed to a microscope slide and treated to preserve the target DNA/RNA and to make the chromosomes visible under a microscope. The slides can be kept in a freezer at 80 °C for at least a year.
Preparation of probes
The probes are single-stranded DNA or RNA sequences complementary to the target sequences. The probes are labeled with a fluorescent dye to make them visible under a fluorescence microscope.
Probe labeling
Label the FISH probes with a fluorescent dye, which will bind to specific regions of the target DNA. Probes can either be directly fluorophore-labeled or indirectly labeled with a hapten that can later be detected via an enzymatic or immunological detection technique.
There are now several techniques for tagging DNA probes for nonradioactive in situ hybridization. The most popular strategy is to label the probe with reporter chemicals (haptens). There are several different haptens on the market, including biotin, digoxigenin, dinitrophenol, fluorescein, rhodamine, AMCA, and coumarin. According to standard protocols, these haptens can be included as labeled nucleotides using the nick translation, random primer labeling, or PCR tagging techniques.
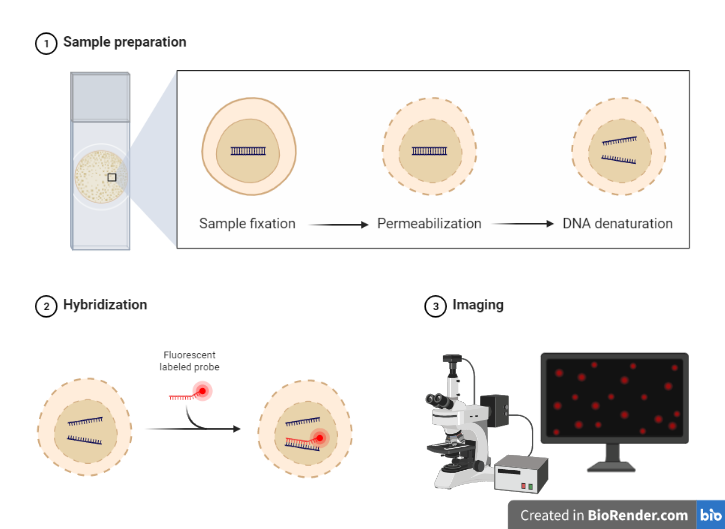
Fig: Steps in FISH Technique
Denaturation: The process of breaking the hydrogen bonds in a double-stranded DNA molecule to create a single-stranded DNA molecule is known as denaturation. Typically, this is accomplished by heating the DNA sample to a high temperature (between 80 and 100 °C), which separates the DNA strands. The complementary probe can then be hybridized with the single-stranded DNA molecules to enable the imaging and localisation of particular DNA sequences within cells or tissues.
Hybridization: Incubate the sample with the labeled probes, allowing them to hybridize (bind) to the target DNA. During hybridization, the labeled probes are added to the sample and are allowed to bind to the complementary DNA sequences in the target. This binding process is specific, meaning that the probes will only bind to their intended target, allowing the researcher to visualize specific regions of the DNA. Hybridization conditions, such as temperature, salt concentration, and incubation time, are carefully controlled to ensure optimal binding of the probes to the target DNA
Washing: Wash the sample to remove unbound probes, reduce background signal, and increase the specificity. A high salt concentration in the washing solution often aids in the removal of unbound probes and the reduction of non-specific binding. Multiple washings of the sample are possible to further drastically reduce background signal.
Detection: Visualize the bound probes using fluorescence microscopy and capture images. Visualization in this technique typically involves the use of fluorescence microscopy to detect and visualize the bound probes. The sample is viewed under a fluorescence microscope equipped with a filter that is specific for the wavelength of light emitted by the fluorescent dye used to label the probes. In the FISH technique, each probe is labeled with a different fluorescence dye, allowing multiple probes to be visualized simultaneously. The fluorescence signals can be viewed in real-time, or captured as images for later analysis.
In addition, the nonfluorescent hapten must first be seen using an enzymatic or immunological detection technique if the probe is indirectly tagged with it.
Analysis: Analyse the captured images to determine the distribution and number of fluorescent signals, which can provide information about genetic changes in the sample.
Interpretation: The results are interpreted and analyzed, taking into account the pattern of fluorescence signals to determine the presence, absence or rearrangement of the target sequences. Interpret the results, which can provide insight into genetic changes, chromosomal aberrations, gene expression patterns, and other biological processes.
Reporting: Report the results, which can be used to guide further research and inform clinical decision making.
Applications:
Prenatal diagnosis of chromosomal abnormalities: It is a widely used technique in cytogenetics to visualize specific DNA sequences in chromosomes and to detect chromosomal abnormalities. FISH can detect specific chromosomal abnormalities, such as deletions, duplications, and translocations, which are often associated with genetic disorders and cancer. In addition, this technique is used in prenatal diagnosis to detect chromosomal abnormalities in a developing foetus, providing important information for genetic counselling and treatment planning.
Detection of copy number variants (CNVs): Copy number variants (CNVs) refer to changes in the number of copies of a specific DNA sequence in a genome. These changes can result in gains or losses of genetic material, which can contribute to genetic disorders and diseases, such as cancer. Fluorescence In Situ Hybridization (FISH) is a widely used technique for the detection of CNVs.
Detection of cancer: FISH is used to diagnose various types of cancer, including leukemia, lymphoma, and solid tumors. The technique can detect specific chromosomal abnormalities and gene rearrangements that are characteristic of certain cancers. It can be used to monitor the response of a cancer to treatment by detecting changes in gene expression and chromosomal structure. This information can help to determine the effectiveness of the treatment and whether a change in treatment is necessary.
Detection of infectious diseases: FISH is used to identify infectious pathogens by analyzing 16S ribosomal RNA (rRNA) that is distinctive to phylogenetic groups. For the investigation of microbial communities in the mouth cavity and gastrointestinal flora, FISH probes made of oligonucleotide sequences (17–34 nucleotides in length) complementary to 16S rRNA can be utilized. The microbial population is highly populated in the intestine and mouth cavity, and FISH has been used to identify pathogens there. The bacteria causing respiratory tract infections have also been targeted in the design of specific oligonucleotide probes. FISH has also been used to identify infections in tissues in a manner similar to that. Pathogenic bacteria in blood cultures have been identified using genus- and species-specific oligonucleotide probes. For instance, FISH probes that are complementary to a certain 16s rRNA sequence can identify malaria infection in blood samples.
Genetics: FISH is used to identify and locate specific genetic mutations in a sample, providing important information for genetic counselling and diagnosis of genetic disorders.
Drug discovery: FISH is used in drug discovery research to evaluate the effectiveness of new drugs by tracking changes in gene expression and chromosomal structure in response to drug treatment.
Neuroscience: FISH is used in neuroscience research to study the localization and expression of specific genes in the brain, providing important insights into the biology of the nervous system.
Limitations:
When using several probes in a single experiment, FISH can be complicated to perform and analyze, requiring specific tools and knowledge. Similar to FISH probes, non-specific targets may bind to them, producing false-positive results and making interpretation more difficult. With an usual resolution of around 10–50 kb, this approach has a relatively low resolution. This implies that it might not be able to recognize tiny CNVs or alterations in the genome’s structural elements.
While FISH is an effective method for identifying particular DNA sequences and chromosomal aberrations, there are some drawbacks that must be taken into account when determining if FISH is the most appropriate method for a given study.
References:
- Henegariu, O., Bray-Ward, P., Artan, S., Vance, G. H., Qumsyieh, M. and Ward, D. C. (2001) ‘Small marker chromosome identification in metaphase and interphase using centromeric multiplex FISH (CM-FISH)’, Laboratory investigation, 81(4), 475-481.
- Parra, I., & Windle, B. High resolution visual mapping of stretched DNA by fluorescent hybridization. Nature Genetics 5, 17–21 (1993)
- Fluorescence In situ Hybridization: Cell-Based Genetic Diagnostic and Research Applications.
- Shakoori AR. Fluorescence In Situ Hybridization (FISH) and Its Applications. Chromosome Structure and Aberrations. 2017 Feb 10:343–67.
